Domain Expansion and Functional Diversification in Vertebrate Reproductive Proteins
- PMID: 35587583
- PMCID: PMC9154058
- DOI: 10.1093/molbev/msac105
Domain Expansion and Functional Diversification in Vertebrate Reproductive Proteins
Abstract
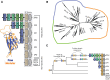
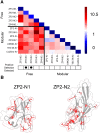
The rapid evolution of fertilization proteins has generated remarkable diversity in molecular structure and function. Glycoproteins of vertebrate egg coats contain multiple zona pellucida (ZP)-N domains (1-6 copies) that facilitate multiple reproductive functions, including species-specific sperm recognition. In this report, we integrate phylogenetics and machine learning to investigate how ZP-N domains diversify in structure and function. The most C-terminal ZP-N domain of each paralog is associated with another domain type (ZP-C), which together form a "ZP module." All modular ZP-N domains are phylogenetically distinct from nonmodular or free ZP-N domains. Machine learning-based classification identifies eight residues that form a stabilizing network in modular ZP-N domains that is absent in free domains. Positive selection is identified in some free ZP-N domains. Our findings support that strong purifying selection has conserved an essential structural core in modular ZP-N domains, with the relaxation of this structural constraint allowing free N-terminal domains to functionally diversify.
Keywords: fertilization; gene duplication; machine learning; molecular evolution; phylogenetics; protein structure.
© The Author(s) 2022. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

Similar articles
-
Isolated ZP-N domains constitute the N-terminal extensions of Zona Pellucida proteins.Bioinformatics. 2007 Aug 1;23(15):1871-4. doi: 10.1093/bioinformatics/btm265. Epub 2007 May 17. Bioinformatics. 2007. PMID: 17510169
-
Egg Coat Proteins Across Metazoan Evolution.Curr Top Dev Biol. 2018;130:443-488. doi: 10.1016/bs.ctdb.2018.03.005. Epub 2018 May 7. Curr Top Dev Biol. 2018. PMID: 29853187 Free PMC article. Review.
-
Proteomic characterization and evolutionary analyses of zona pellucida domain-containing proteins in the egg coat of the cephalochordate, Branchiostoma belcheri.BMC Evol Biol. 2012 Dec 8;12:239. doi: 10.1186/1471-2148-12-239. BMC Evol Biol. 2012. PMID: 23216630 Free PMC article.
-
Zona pellucida domain proteins.Annu Rev Biochem. 2005;74:83-114. doi: 10.1146/annurev.biochem.74.082803.133039. Annu Rev Biochem. 2005. PMID: 15952882 Review.
-
Molecular Evolutionary Analysis of Nematode Zona Pellucida (ZP) Modules Reveals Disulfide-Bond Reshuffling and Standalone ZP-C Domains.Genome Biol Evol. 2020 Aug 1;12(8):1240-1255. doi: 10.1093/gbe/evaa095. Genome Biol Evol. 2020. PMID: 32426804 Free PMC article.
Cited by
-
Pollen Coat Proteomes of Arabidopsis thaliana, Arabidopsis lyrata, and Brassica oleracea Reveal Remarkable Diversity of Small Cysteine-Rich Proteins at the Pollen-Stigma Interface.Biomolecules. 2023 Jan 12;13(1):157. doi: 10.3390/biom13010157. Biomolecules. 2023. PMID: 36671543 Free PMC article.
References
-
- Anisimova M, Liberles D. 2012. Detecting and understanding natural selection. In: Cannarozzi G, Schneider A, editors. Codon evolution mechanisms and models. Oxford: Oxford University Press.
-
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodol). 57(1):289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

