Transcriptome analysis reveals high tumor heterogeneity with respect to re-activation of stemness and proliferation programs
- PMID: 35587924
- PMCID: PMC9119523
- DOI: 10.1371/journal.pone.0268626
Transcriptome analysis reveals high tumor heterogeneity with respect to re-activation of stemness and proliferation programs
Abstract
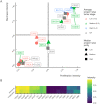
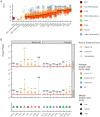
Significant alterations in signaling pathways and transcriptional regulatory programs together represent major hallmarks of many cancers. These, among all, include the reactivation of stemness, which is registered by the expression of pathways that are active in the embryonic stem cells (ESCs). Here, we assembled gene sets that reflect the stemness and proliferation signatures and used them to analyze a large panel of RNA-seq data from The Cancer Genome Atlas (TCGA) Consortium in order to specifically assess the expression of stemness-related and proliferation-related genes across a collection of different tumor types. We introduced a metric that captures the collective similarity of the expression profile of a tumor to that of ESCs, which showed that stemness and proliferation signatures vary greatly between different tumor types. We also observed a high degree of intertumoral heterogeneity in the expression of stemness- and proliferation-related genes, which was associated with increased hazard ratios in a fraction of tumors and mirrored by high intratumoral heterogeneity and a remarkable stemness capacity in metastatic lesions across cancer cells in single cell RNA-seq datasets. Taken together, these results indicate that the expression of stemness signatures is highly heterogeneous and cannot be used as a universal determinant of cancer. This calls into question the universal validity of diagnostic tests that are based on stem cell markers.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Roth GA, Abate D, Abate KHea. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392(10159):1736–1788. doi: 10.1016/S0140-6736(18)32203-7 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

