A crowd of BashTheBug volunteers reproducibly and accurately measure the minimum inhibitory concentrations of 13 antitubercular drugs from photographs of 96-well broth microdilution plates
- PMID: 35588296
- PMCID: PMC9286738
- DOI: 10.7554/eLife.75046
A crowd of BashTheBug volunteers reproducibly and accurately measure the minimum inhibitory concentrations of 13 antitubercular drugs from photographs of 96-well broth microdilution plates
Abstract
Tuberculosis is a respiratory disease that is treatable with antibiotics. An increasing prevalence of resistance means that to ensure a good treatment outcome it is desirable to test the susceptibility of each infection to different antibiotics. Conventionally, this is done by culturing a clinical sample and then exposing aliquots to a panel of antibiotics, each being present at a pre-determined concentration, thereby determining if the sample isresistant or susceptible to each sample. The minimum inhibitory concentration (MIC) of a drug is the lowestconcentration that inhibits growth and is a more useful quantity but requires each sample to be tested at a range ofconcentrations for each drug. Using 96-well broth micro dilution plates with each well containing a lyophilised pre-determined amount of an antibiotic is a convenient and cost-effective way to measure the MICs of several drugs at once for a clinical sample. Although accurate, this is still an expensive and slow process that requires highly-skilled and experienced laboratory scientists. Here we show that, through the BashTheBug project hosted on the Zooniverse citizen science platform, a crowd of volunteers can reproducibly and accurately determine the MICs for 13 drugs and that simply taking the median or mode of 11-17 independent classifications is sufficient. There is therefore a potential role for crowds to support (but not supplant) the role of experts in antibiotic susceptibility testing.
Keywords: M. tuberculosis; antibiotics; citizen science; clinical microbiology; infectious disease; microbiology; tuberculosis.
Plain language summary
Tuberculosis is a bacterial respiratory infection that kills about 1.4 million people worldwide each year. While antibiotics can cure the condition, the bacterium responsible for this disease, Mycobacterium tuberculosis, is developing resistance to these treatments. Choosing which antibiotics to use to treat the infection more carefully may help to combat the growing threat of drug-resistant bacteria. One way to find the best choice is to test how an antibiotic affects the growth of M. tuberculosis in the laboratory. To speed up this process, laboratories test multiple drugs simultaneously. They do this by growing bacteria on plates with 96 wells and injecting individual antibiotics in to each well at different concentrations. The Comprehensive Resistance Prediction for Tuberculosis (CRyPTIC) consortium has used this approach to collect and analyse bacteria from over 20,000 tuberculosis patients. An image of the 96-well plate is then captured and the level of bacterial growth in each well is assessed by laboratory scientists. But this work is difficult, time-consuming, and subjective, even for tuberculosis experts. Here, Fowler et al. show that enlisting citizen scientists may help speed up this process and reduce errors that arise from analysing such a large dataset. In April 2017, Fowler et al. launched the project ‘BashTheBug’ on the Zooniverse citizen science platform where anyone can access and analyse the images from the CRyPTIC consortium. They found that a crowd of inexperienced volunteers were able to consistently and accurately measure the concentration of antibiotics necessary to inhibit the growth of M. tuberculosis. If the concentration is above a pre-defined threshold, the bacteria are considered to be resistant to the treatment. A consensus result could be reached by calculating the median value of the classifications provided by as few as 17 different BashTheBug participants. The work of BashTheBug volunteers has reduced errors in the CRyPTIC project data, which has been used for several other studies. For instance, the World Health Organization (WHO) has also used the data to create a catalogue of genetic mutations associated with antibiotics resistance in M. tuberculosis. Enlisting citizen scientists has accelerated research on tuberculosis and may help with other pressing public health concerns.
© 2022, Fowler et al.
Conflict of interest statement
PF, CW, HS, TZ, EB, SH, AG, AR, SK, TW, TP, GM, CL, DC, DC, AW No competing interests declared
Figures
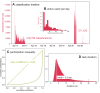












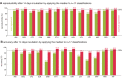
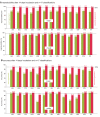


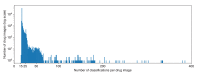



References
-
- Cox J, Oh EY, Simmons B, Lintott C, Masters K, Greenhill A, Graham G, Holmes K. Defining and Measuring Success in Online Citizen Science: A Case Study of Zooniverse Projects. Computing in Science & Engineering. 2015;17:28–41. doi: 10.1109/MCSE.2015.65. - DOI
-
- Fowler PW. Help us fight antibiotic resistance. 2017. [June 10, 2022]. https://bashthebug.net/bashthebug-on-the-zooniverse/
-
- Fowler PW. pyniverse: a Python package to analyse classifications made by volunteers in a generic Zooniverse citizen science project. ce066f5Github. 2018a https://github.com/fowler-lab/pyniverse
-
- Fowler PW. bashthebug: a Python package to analyse the results of the Zooniverse volunteers for the BashTheBug citizen science project. 8b22907Github. 2018b https://github.com/fowler-lab/bashthebug
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

