Caspase-4/11 exacerbates disease severity in SARS-CoV-2 infection by promoting inflammation and immunothrombosis
- PMID: 35588457
- PMCID: PMC9173818
- DOI: 10.1073/pnas.2202012119
Caspase-4/11 exacerbates disease severity in SARS-CoV-2 infection by promoting inflammation and immunothrombosis
Abstract
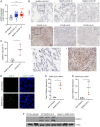
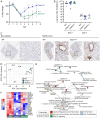
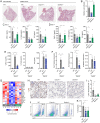
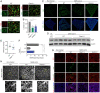
Severe acute respiratory syndrome coronavirus 2 (SARS–CoV-2) is a worldwide health concern, and new treatment strategies are needed. Targeting inflammatory innate immunity pathways holds therapeutic promise, but effective molecular targets remain elusive. Here, we show that human caspase-4 (CASP4) and its mouse homolog, caspase-11 (CASP11), are up-regulated in SARS–CoV-2 infections and that CASP4 expression correlates with severity of SARS–CoV-2 infection in humans. SARS–CoV-2–infected Casp11−/− mice were protected from severe weight loss and lung pathology, including blood vessel damage, compared to wild-type (WT) mice and mice lacking the caspase downstream effector gasdermin-D (Gsdmd−/−). Notably, viral titers were similar regardless of CASP11 knockout. Global transcriptomics of SARS–CoV-2–infected WT, Casp11−/−, and Gsdmd−/− lungs identified restrained expression of inflammatory molecules and altered neutrophil gene signatures in Casp11−/− mice. We confirmed that protein levels of inflammatory mediators interleukin (IL)-1β, IL-6, and CXCL1, as well as neutrophil functions, were reduced in Casp11−/− lungs. Additionally, Casp11−/− lungs accumulated less von Willebrand factor, a marker for endothelial damage, but expressed more Kruppel-Like Factor 2, a transcription factor that maintains vascular integrity. Overall, our results demonstrate that CASP4/11 promotes detrimental SARS–CoV-2–induced inflammation and coagulopathy, largely independently of GSDMD, identifying CASP4/11 as a promising drug target for treatment and prevention of severe COVID-19.
Keywords: SARS–CoV-2; innate immunity; thrombosis.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
- R21 AG064899/AG/NIA NIH HHS/United States
- R21 AI142256/AI/NIAID NIH HHS/United States
- P51 OD011132/OD/NIH HHS/United States
- R56 AI157872/AI/NIAID NIH HHS/United States
- R01 AI123661/AI/NIAID NIH HHS/United States
- R21 AI151230/AI/NIAID NIH HHS/United States
- R01 AI157205/AI/NIAID NIH HHS/United States
- T32 AI074492/AI/NIAID NIH HHS/United States
- R01 DK101323/DK/NIDDK NIH HHS/United States
- R01 AI145144/AI/NIAID NIH HHS/United States
- R01 HL154001/HL/NHLBI NIH HHS/United States
- R01 HL159675/HL/NHLBI NIH HHS/United States
- R21 AG067755/AG/NIA NIH HHS/United States
- R01 AI124121/AI/NIAID NIH HHS/United States
- R21 AI163753/AI/NIAID NIH HHS/United States
- R01 AI159452/AI/NIAID NIH HHS/United States
- U54 CA260582/CA/NCI NIH HHS/United States
- HHSN272201400004C/AI/NIAID NIH HHS/United States
- K22 AI146141/AI/NIAID NIH HHS/United States
- T32 AI165391/AI/NIAID NIH HHS/United States
- R01 HL127651/HL/NHLBI NIH HHS/United States
- R01 AI130110/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

