Identifying who has long COVID in the USA: a machine learning approach using N3C data
- PMID: 35589549
- PMCID: PMC9110014
- DOI: 10.1016/S2589-7500(22)00048-6
Identifying who has long COVID in the USA: a machine learning approach using N3C data
Abstract
Background: Post-acute sequelae of SARS-CoV-2 infection, known as long COVID, have severely affected recovery from the COVID-19 pandemic for patients and society alike. Long COVID is characterised by evolving, heterogeneous symptoms, making it challenging to derive an unambiguous definition. Studies of electronic health records are a crucial element of the US National Institutes of Health's RECOVER Initiative, which is addressing the urgent need to understand long COVID, identify treatments, and accurately identify who has it-the latter is the aim of this study.
Methods: Using the National COVID Cohort Collaborative's (N3C) electronic health record repository, we developed XGBoost machine learning models to identify potential patients with long COVID. We defined our base population (n=1 793 604) as any non-deceased adult patient (age ≥18 years) with either an International Classification of Diseases-10-Clinical Modification COVID-19 diagnosis code (U07.1) from an inpatient or emergency visit, or a positive SARS-CoV-2 PCR or antigen test, and for whom at least 90 days have passed since COVID-19 index date. We examined demographics, health-care utilisation, diagnoses, and medications for 97 995 adults with COVID-19. We used data on these features and 597 patients from a long COVID clinic to train three machine learning models to identify potential long COVID among all patients with COVID-19, patients hospitalised with COVID-19, and patients who had COVID-19 but were not hospitalised. Feature importance was determined via Shapley values. We further validated the models on data from a fourth site.
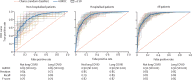
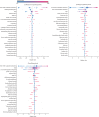
Findings: Our models identified, with high accuracy, patients who potentially have long COVID, achieving areas under the receiver operator characteristic curve of 0·92 (all patients), 0·90 (hospitalised), and 0·85 (non-hospitalised). Important features, as defined by Shapley values, include rate of health-care utilisation, patient age, dyspnoea, and other diagnosis and medication information available within the electronic health record.
Interpretation: Patients identified by our models as potentially having long COVID can be interpreted as patients warranting care at a specialty clinic for long COVID, which is an essential proxy for long COVID diagnosis as its definition continues to evolve. We also achieve the urgent goal of identifying potential long COVID in patients for clinical trials. As more data sources are identified, our models can be retrained and tuned based on the needs of individual studies.
Funding: US National Institutes of Health and National Center for Advancing Translational Sciences through the RECOVER Initiative.
Copyright © 2022 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY-NC-ND 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests ATG is an employee of Palantir Technologies. ERP, JPD, SEJ, RRD, CGC, TDB, JAM, RM, AW, and MAH report research funding from the NIH. ERP and MGK report research funding from PCORI. MAH and JAM are co-founders of Pryzm Health. All other authors declare no competing interests.
Figures



References
-
- Greenhalgh T, Knight M, A'Court C, Buxton M, Husain L. Management of post-acute covid-19 in primary care. BMJ. 2020;370 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

