Integrating single-cell RNA sequencing with spatial transcriptomics reveals immune landscape for interstitial cystitis
- PMID: 35589692
- PMCID: PMC9120182
- DOI: 10.1038/s41392-022-00962-8
Integrating single-cell RNA sequencing with spatial transcriptomics reveals immune landscape for interstitial cystitis
Abstract
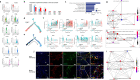
Interstitial cystitis (IC) is a severely debilitating and chronic disorder with unclear etiology and pathophysiology, which makes the diagnosis difficult and treatment challenging. To investigate the role of immunity in IC bladders, we sequenced 135,091 CD45+ immune cells from 15 female patients with IC and 9 controls with stress urinary incontinence using single-cell RNA sequencing (scRNA-seq). 22 immune subpopulations were identified in the constructed landscape. Among them, M2-like macrophages, inflammatory CD14+ macrophages, and conventional dendritic cells had the most communications with other immune cells. Then, a significant increase of central memory CD4+ T cells, regulatory T cells, GZMK+CD8+ T cells, activated B cells, un-switched memory B cells, and neutrophils, and a significant decrease of CD8+ effector T cells, Th17 cells, follicular helper T cells, switched memory B cells, transitional B cells, and macrophages were noted in IC bladders. The enrichment analysis identified a virus-related response during the dynamic change of cell proportion, furthermore, the human polyomavirus-2 was detected with a positive rate of 95% in urine of patients with IC. By integrating the results of scRNA-seq with spatial transcriptomics, we found nearly all immune subpopulations were enriched in the urothelial region or located close to fibroblasts in IC bladders, but they were discovered around urothelium and smooth muscle cells in control bladders. These findings depict the immune landscape for IC and might provide valuable insights into the pathophysiology of IC.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

