BMP4 drives primed to naïve transition through PGC-like state
- PMID: 35589713
- PMCID: PMC9120449
- DOI: 10.1038/s41467-022-30325-4
BMP4 drives primed to naïve transition through PGC-like state
Abstract
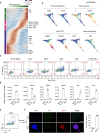
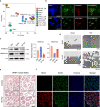
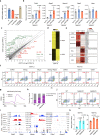
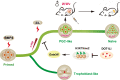
Multiple pluripotent states have been described in mouse and human stem cells. Here, we apply single-cell RNA-seq to a newly established BMP4 induced mouse primed to naïve transition (BiPNT) system and show that the reset is not a direct reversal of cell fate but goes through a primordial germ cell-like cells (PGCLCs) state. We first show that epiblast stem cells bifurcate into c-Kit+ naïve and c-Kit- trophoblast-like cells, among which, the naïve branch undergoes further transition through a PGCLCs intermediate capable of spermatogenesis in vivo. Mechanistically, we show that DOT1L inhibition permits the transition from primed pluripotency to PGCLCs in part by facilitating the loss of H3K79me2 from Gata3/6. In addition, Prdm1/Blimp1 is required for PGCLCs and naïve cells, while Gata2 inhibits PGC-like state by promoting trophoblast-like fate. Our work not only reveals an alternative route for primed to naïve transition, but also gains insight into germ cell development.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
NANOG alone induces germ cells in primed epiblast in vitro by activation of enhancers.Nature. 2016 Jan 21;529(7586):403-407. doi: 10.1038/nature16480. Epub 2016 Jan 11. Nature. 2016. PMID: 26751055 Free PMC article.
-
Production and Analysis of Human Primordial Germ Cell-Like Cells.Methods Mol Biol. 2021;2195:125-145. doi: 10.1007/978-1-0716-0860-9_10. Methods Mol Biol. 2021. PMID: 32852762 Free PMC article.
-
Stability of Imprinting and Differentiation Capacity in Naïve Human Cells Induced by Chemical Inhibition of CDK8 and CDK19.Cells. 2021 Apr 12;10(4):876. doi: 10.3390/cells10040876. Cells. 2021. PMID: 33921436 Free PMC article.
-
The role of BMP4 signaling in trophoblast emergence from pluripotency.Cell Mol Life Sci. 2022 Jul 25;79(8):447. doi: 10.1007/s00018-022-04478-w. Cell Mol Life Sci. 2022. PMID: 35877048 Free PMC article. Review.
-
The post-inner cell mass intermediate: implications for stem cell biology and assisted reproductive technology.Hum Reprod Update. 2015 Sep-Oct;21(5):616-26. doi: 10.1093/humupd/dmv028. Epub 2015 Jun 18. Hum Reprod Update. 2015. PMID: 26089403 Review.
Cited by
-
Spatial transcriptomic characterization of a Carnegie stage 7 human embryo.Nat Cell Biol. 2025 Feb;27(2):360-369. doi: 10.1038/s41556-024-01597-3. Epub 2025 Jan 10. Nat Cell Biol. 2025. PMID: 39794460
-
BRD8 Guards the Pluripotent State by Sensing and Maintaining Histone Acetylation.Adv Sci (Weinh). 2025 Feb;12(5):e2409160. doi: 10.1002/advs.202409160. Epub 2024 Dec 10. Adv Sci (Weinh). 2025. PMID: 39656858 Free PMC article.
-
The SWI/SNF ATP-dependent chromatin remodeling complex in cell lineage priming and early development.Biochem Soc Trans. 2024 Apr 24;52(2):603-616. doi: 10.1042/BST20230416. Biochem Soc Trans. 2024. PMID: 38572912 Free PMC article. Review.
-
Effect of iodoacetic acid on the reproductive system of male mice.Front Pharmacol. 2022 Aug 26;13:958204. doi: 10.3389/fphar.2022.958204. eCollection 2022. Front Pharmacol. 2022. PMID: 36091762 Free PMC article.
-
Cpt1a Drives primed-to-naïve pluripotency transition through lipid remodeling.Commun Biol. 2024 Sep 30;7(1):1223. doi: 10.1038/s42003-024-06874-3. Commun Biol. 2024. PMID: 39349670 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

