Structural insights into the mechanism of pancreatic KATP channel regulation by nucleotides
- PMID: 35589716
- PMCID: PMC9120461
- DOI: 10.1038/s41467-022-30430-4
Structural insights into the mechanism of pancreatic KATP channel regulation by nucleotides
Abstract
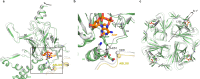
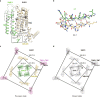
ATP-sensitive potassium channels (KATP) are metabolic sensors that convert the intracellular ATP/ADP ratio to the excitability of cells. They are involved in many physiological processes and implicated in several human diseases. Here we present the cryo-EM structures of the pancreatic KATP channel in both the closed state and the pre-open state, resolved in the same sample. We observe the binding of nucleotides at the inhibitory sites of the Kir6.2 channel in the closed but not in the pre-open state. Structural comparisons reveal the mechanism for ATP inhibition and Mg-ADP activation, two fundamental properties of KATP channels. Moreover, the structures also uncover the activation mechanism of diazoxide-type KATP openers.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

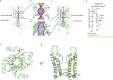

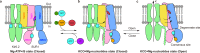
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

