Noninvasive urinary protein signatures associated with colorectal cancer diagnosis and metastasis
- PMID: 35589723
- PMCID: PMC9119985
- DOI: 10.1038/s41467-022-30391-8
Noninvasive urinary protein signatures associated with colorectal cancer diagnosis and metastasis
Abstract
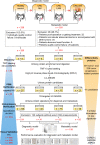
Currently, imaging, fecal immunochemical tests (FITs) and serum carcinoembryonic antigen (CEA) tests are not adequate for the early detection and evaluation of metastasis and recurrence in colorectal cancer (CRC). To comprehensively identify and validate more accurate noninvasive biomarkers in urine, we implement a staged discovery-verification-validation pipeline in 657 urine and 993 tissue samples from healthy controls and CRC patients with a distinct metastatic risk. The generated diagnostic signature combined with the FIT test reveals a significantly increased sensitivity (+21.2% in the training set, +43.7% in the validation set) compared to FIT alone. Moreover, the generated metastatic signature for risk stratification correctly predicts over 50% of CEA-negative metastatic patients. The tissue validation shows that elevated urinary protein biomarkers reflect their alterations in tissue. Here, we show promising urinary protein signatures and provide potential interventional targets to reliably detect CRC, although further multi-center external validation is needed to generalize the findings.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Pioneering noninvasive colorectal cancer detection with an AI-enhanced breath volatilomics platform.Theranostics. 2024 Jul 8;14(11):4240-4255. doi: 10.7150/thno.94950. eCollection 2024. Theranostics. 2024. PMID: 39113791 Free PMC article.
-
Identification and Validation of MicroRNA Profiles in Fecal Samples for Detection of Colorectal Cancer.Gastroenterology. 2020 Mar;158(4):947-957.e4. doi: 10.1053/j.gastro.2019.10.005. Epub 2019 Oct 14. Gastroenterology. 2020. PMID: 31622624
-
Fecal Clostridium symbiosum for Noninvasive Detection of Early and Advanced Colorectal Cancer: Test and Validation Studies.EBioMedicine. 2017 Nov;25:32-40. doi: 10.1016/j.ebiom.2017.10.005. Epub 2017 Oct 4. EBioMedicine. 2017. PMID: 29033369 Free PMC article. Clinical Trial.
-
Carcinoembryonic Antigen (CEA) and Hepatic Metastasis in Colorectal Cancer: Update on Biomarker for Clinical and Biotechnological Approaches.Recent Pat Biotechnol. 2018;12(4):269-279. doi: 10.2174/1872208312666180731104244. Recent Pat Biotechnol. 2018. PMID: 30062978 Review.
-
Methylated Septin 9 as a Promising Biomarker in the Diagnosis and Recurrence Monitoring of Colorectal Cancer.Dis Markers. 2022 Jul 2;2022:7087885. doi: 10.1155/2022/7087885. eCollection 2022. Dis Markers. 2022. PMID: 35818587 Free PMC article. Review.
Cited by
-
Frontiers in mass spectrometry-based clinical proteomics for cancer diagnosis and treatment.Cancer Sci. 2023 May;114(5):1783-1791. doi: 10.1111/cas.15731. Epub 2023 Feb 1. Cancer Sci. 2023. PMID: 36661476 Free PMC article. Review.
-
A simple urine test by 3D-plus-3D immunoassay guides precise in vitro cancer diagnosis.Bioeng Transl Med. 2023 Jan 18;8(3):e10489. doi: 10.1002/btm2.10489. eCollection 2023 May. Bioeng Transl Med. 2023. PMID: 37206218 Free PMC article.
-
Machine learning model for predicting age in healthy individuals using age-related gut microbes and urine metabolites.Gut Microbes. 2023 Jan-Dec;15(1):2226915. doi: 10.1080/19490976.2023.2226915. Gut Microbes. 2023. PMID: 37351626 Free PMC article.
-
Proteomics-driven noninvasive screening of circulating serum protein panels for the early diagnosis of hepatocellular carcinoma.Nat Commun. 2023 Dec 18;14(1):8392. doi: 10.1038/s41467-023-44255-2. Nat Commun. 2023. PMID: 38110372 Free PMC article.
-
Urinary complement factor D is increased in primary malignant hypertension: a single-center, cross-sectional study.Sci Rep. 2024 Jul 15;14(1):16253. doi: 10.1038/s41598-024-66875-4. Sci Rep. 2024. PMID: 39009768 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

