Characterizing the mucin-degrading capacity of the human gut microbiota
- PMID: 35589783
- PMCID: PMC9120202
- DOI: 10.1038/s41598-022-11819-z
Characterizing the mucin-degrading capacity of the human gut microbiota
Abstract
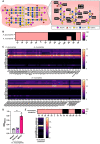
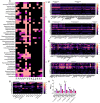
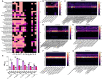
Mucin-degrading microbes are known to harbor glycosyl hydrolases (GHs) which cleave specific glycan linkages. Although several microbial species have been identified as mucin degraders, there are likely many other members of the healthy gut community with the capacity to degrade mucins. The aim of the present study was to systematically examine the CAZyme mucin-degrading profiles of the human gut microbiota. Within the Verrucomicrobia phylum, all Akkermansia glycaniphila and muciniphila genomes harbored multiple gene copies of mucin-degrading GHs. The only representative of the Lentisphaerae phylum, Victivallales, harbored a GH profile that closely mirrored Akkermansia. In the Actinobacteria phylum, we found several Actinomadura, Actinomyces, Bifidobacterium, Streptacidiphilus and Streptomyces species with mucin-degrading GHs. Within the Bacteroidetes phylum, Alistipes, Alloprevotella, Bacteroides, Fermenitomonas Parabacteroides, Prevotella and Phocaeicola species had mucin degrading GHs. Firmicutes contained Abiotrophia, Blautia, Enterococcus, Paenibacillus, Ruminococcus, Streptococcus, and Viridibacillus species with mucin-degrading GHs. Interestingly, far fewer mucin-degrading GHs were observed in the Proteobacteria phylum and were found in Klebsiella, Mixta, Serratia and Enterobacter species. We confirmed the mucin-degrading capability of 23 representative gut microbes using a chemically defined media lacking glucose supplemented with porcine intestinal mucus. These data greatly expand our knowledge of microbial-mediated mucin degradation within the human gut microbiota.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Corfield AP, Wagner SA, Clamp JR, Kriaris MS, Hoskins LC. Mucin degradation in the human colon: Production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect. Immun. 1992;60:3971–3978. doi: 10.1128/iai.60.10.3971-3978.1992. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

