Rituximab versus tocilizumab in rheumatoid arthritis: synovial biopsy-based biomarker analysis of the phase 4 R4RA randomized trial
- PMID: 35589854
- PMCID: PMC9205785
- DOI: 10.1038/s41591-022-01789-0
Rituximab versus tocilizumab in rheumatoid arthritis: synovial biopsy-based biomarker analysis of the phase 4 R4RA randomized trial
Abstract
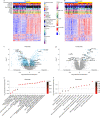
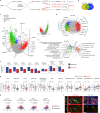
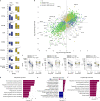
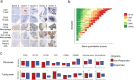
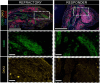
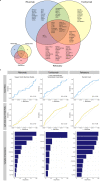
Patients with rheumatoid arthritis (RA) receive highly targeted biologic therapies without previous knowledge of target expression levels in the diseased tissue. Approximately 40% of patients do not respond to individual biologic therapies and 5-20% are refractory to all. In a biopsy-based, precision-medicine, randomized clinical trial in RA (R4RA; n = 164), patients with low/absent synovial B cell molecular signature had a lower response to rituximab (anti-CD20 monoclonal antibody) compared with that to tocilizumab (anti-IL6R monoclonal antibody) although the exact mechanisms of response/nonresponse remain to be established. Here, in-depth histological/molecular analyses of R4RA synovial biopsies identify humoral immune response gene signatures associated with response to rituximab and tocilizumab, and a stromal/fibroblast signature in patients refractory to all medications. Post-treatment changes in synovial gene expression and cell infiltration highlighted divergent effects of rituximab and tocilizumab relating to differing response/nonresponse mechanisms. Using ten-by-tenfold nested cross-validation, we developed machine learning algorithms predictive of response to rituximab (area under the curve (AUC) = 0.74), tocilizumab (AUC = 0.68) and, notably, multidrug resistance (AUC = 0.69). This study supports the notion that disease endotypes, driven by diverse molecular pathology pathways in the diseased tissue, determine diverse clinical and treatment-response phenotypes. It also highlights the importance of integration of molecular pathology signatures into clinical algorithms to optimize the future use of existing medications and inform the development of new drugs for refractory patients.
© 2022. Crown.
Conflict of interest statement
S.E.C., B.M.H. and S.E.W. are employees and stockholders of NanoString Technologies, Inc. C.P. and M.J.L. are inventors on a patent application (no. GB 2100821.4), submitted by Queen Mary University of London, that covers methods used to select treatments in RA. All other authors declare no competing interests.
Figures






Comment in
-
Toward pre-treatment prediction of biologic DMARD response in RA.Nat Rev Rheumatol. 2022 Jul;18(7):365. doi: 10.1038/s41584-022-00800-9. Nat Rev Rheumatol. 2022. PMID: 35672462 No abstract available.
-
Inching closer to precision treatment for rheumatoid arthritis.Nat Med. 2022 Jun;28(6):1129-1131. doi: 10.1038/s41591-022-01857-5. Nat Med. 2022. PMID: 35681106 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

