Localization of EccA3 at the growing pole in Mycobacterium smegmatis
- PMID: 35590245
- PMCID: PMC9118679
- DOI: 10.1186/s12866-022-02554-6
Localization of EccA3 at the growing pole in Mycobacterium smegmatis
Abstract
Background: Bacteria require specialized secretion systems for the export of molecules into the extracellular space to modify their environment and scavenge for nutrients. The ESX-3 secretion system is required by mycobacteria for iron homeostasis. The ESX-3 operon encodes for one cytoplasmic component (EccA3) and five membrane components (EccB3 - EccE3 and MycP3). In this study we sought to identify the sub-cellular location of EccA3 of the ESX-3 secretion system in mycobacteria.
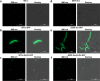
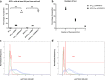
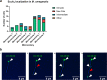
Results: Fluorescently tagged EccA3 localized to a single pole in the majority of Mycobacterium smegmatis cells and time-lapse fluorescent microscopy identified this pole as the growing pole. Deletion of ESX-3 did not prevent polar localization of fluorescently tagged EccA3, suggesting that EccA3 unipolar localization is independent of other ESX-3 components. Affinity purification - mass spectrometry was used to identify EccA3 associated proteins which may contribute to the localization of EccA3 at the growing pole. EccA3 co-purified with fatty acid metabolism proteins (FAS, FadA3, KasA and KasB), mycolic acid synthesis proteins (UmaA, CmaA1), cell division proteins (FtsE and FtsZ), and cell shape and cell cycle proteins (MurS, CwsA and Wag31). Secretion system related proteins Ffh, SecA1, EccA1, and EspI were also identified.
Conclusions: Time-lapse microscopy demonstrated that EccA3 is located at the growing pole in M. smegmatis. The co-purification of EccA3 with proteins known to be required for polar growth, mycolic acid synthesis, the Sec secretion system (SecA1), and the signal recognition particle pathway (Ffh) also suggests that EccA3 is located at the site of active cell growth.
Keywords: ESX-3; EccA3; Mycobacterium; Polar localization; Type VII secretion.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Renshaw PS, Panagiotidou P, Whelan A, Gordon SV, Glyn Hewinson R, Williamson RA, et al. Conclusive evidence that the major T-cell antigens of the Mycobacterium tuberculosis complex ESAT-6 and CFP-10 form a tight, 1:1 complex and characterization of the structural properties of ESAT-6, CFP-10, and the ESAT-6·CFP-10 complex. Implications for p. J Biol Chem. 2002;277(24):21598–21603. doi: 10.1074/jbc.M201625200. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

