Microbiomes of the Sydney Rock Oyster are acquired through both vertical and horizontal transmission
- PMID: 35590396
- PMCID: PMC9118846
- DOI: 10.1186/s42523-022-00186-9
Microbiomes of the Sydney Rock Oyster are acquired through both vertical and horizontal transmission
Abstract
Background: The term holobiont is widely accepted to describe animal hosts and their associated microorganisms. The genomes of all that the holobiont encompasses, are termed the hologenome and it has been proposed as a unit of selection in evolution. To demonstrate that natural selection acts on the hologenome, a significant portion of the associated microbial genomes should be transferred between generations. Using the Sydney Rock Oyster (Saccostrea glomerata) as a model, we tested if the microbes of this broadcast spawning species could be passed down to the next generation by conducting single parent crosses and tracking the microbiome from parent to offspring and throughout early larval stages using 16S rRNA gene amplicon sequencing. From each cross, we sampled adult tissues (mantle, gill, stomach, gonad, eggs or sperm), larvae (D-veliger, umbo, eyed pediveliger, and spat), and the surrounding environment (water and algae feed) for microbial community analysis.
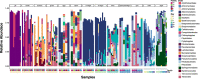
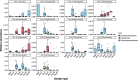
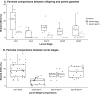
Results: We found that each larval stage has a distinct microbiome that is partially influenced by their parental microbiome, particularly the maternal egg microbiome. We also demonstrate the presence of core microbes that are consistent across all families, persist throughout early life stages (from eggs to spat), and are not detected in the microbiomes of the surrounding environment. In addition to the core microbiomes that span all life cycle stages, there is also evidence of environmentally acquired microbial communities, with earlier larval stages (D-veliger and umbo), more influenced by seawater microbiomes, and later larval stages (eyed pediveliger and spat) dominated by microbial members that are specific to oysters and not detected in the surrounding environment.
Conclusion: Our study characterized the succession of oyster larvae microbiomes from gametes to spat and tracked selected members that persisted across multiple life stages. Overall our findings suggest that both horizontal and vertical transmission routes are possible for the complex microbial communities associated with a broadcast spawning marine invertebrate. We demonstrate that not all members of oyster-associated microbiomes are governed by the same ecological dynamics, which is critical for determining what constitutes a hologenome.
Keywords: Animal microbiomes; Holobiont; Horizontal transmission; Microbiome transmission; Oyster larvae microbiomes; Sydney Rock Oyster; Symbiosis; Vertical transmission.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Gordon J, Knowlton N, Relman DA, Rohwer F, Youle M. Superorganisms and holobionts. Microbe Mag. 2013;8:152–153. doi: 10.1128/microbe.8.152.1. - DOI
-
- Rohwer F, Seguritan V, Azam F, Knowlton N. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser. 2002;243:1–10. doi: 10.3354/meps243001. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
