Genomic Landscape of Actionable Mutations in Primary and Metastatic Tissues of Colon Adenocarcinoma
- PMID: 35592200
- PMCID: PMC9110093
- DOI: 10.7759/cureus.24175
Genomic Landscape of Actionable Mutations in Primary and Metastatic Tissues of Colon Adenocarcinoma
Abstract
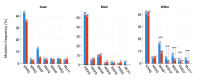
Aim To assess the actionable genomic landscape of colon adenocarcinoma in the primary and metastatic tumor tissues. Methods The data from the American Association for Cancer Research (AACR) Project Genomics Evidence Neoplasia Information Exchange (GENIE) were used in this study. Colon adenocarcinoma patients with primary and metastatic tissue samples (distant organ and lymph node) were selected. Patients with samples from a local recurrence, not otherwise specified tumor samples, and data not collected for sampling localization were excluded. Results A total of 3286 and 1727 patients were included in the primary and metastatic tissue sample groups, respectively. There was no difference between the groups in Kirsten rat sarcoma viral oncogene homolog (KRAS) mutation rates. The rates of v-Raf murine sarcoma viral oncogene homolog B (BRAF) and mismatch repair (MMR) gene mutations were higher in the primary tumor tissues than in the metastatic tumor tissues. There was also no difference between the groups in other actionable gene alterations (e.g. ERBB2 amplification and neurotrophic receptor tyrosine kinase (NTRK) 1 and NTRK3 fusions). In contrast to all cohorts, in Asian and black patients, there was no difference in actionable genomic landscape between the primary and metastatic tumor tissues. Conclusion This study had the largest number of colon cancer patients that evaluated the actionable genomic alterations in primary and metastatic tumor tissues. BRAF and MMR gene alterations were more frequent in the primary tumor tissues than the metastatic tumor tissues.
Keywords: braf mutation; colorectal neoplasms; genomics; k-ras mutations; neoplasm metastasis.
Copyright © 2022, Yekedüz et al.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes.Gastroenterology. 2015 Jan;148(1):88-99. doi: 10.1053/j.gastro.2014.09.041. Epub 2014 Oct 8. Gastroenterology. 2015. PMID: 25305506 Free PMC article. Clinical Trial.
-
Genetic and epigenetic alterations in primary colorectal cancers and related lymph node and liver metastases.Cancer. 2013 Jan 15;119(2):266-76. doi: 10.1002/cncr.27722. Epub 2012 Jul 11. Cancer. 2013. PMID: 22786759
-
The potential of PIK3CA, KRAS, BRAF, and APC hotspot mutations as a non-invasive detection method for colorectal cancer.Mol Cell Probes. 2022 Jun;63:101807. doi: 10.1016/j.mcp.2022.101807. Epub 2022 Mar 13. Mol Cell Probes. 2022. PMID: 35296442
-
Effect of KRAS and BRAF mutations in metastatic colorectal cancer patients: A systematic review and meta-analysis based on tumor sidedness and KRAS subtypes.Hum Antibodies. 2021;29(4):275-284. doi: 10.3233/HAB-210451. Hum Antibodies. 2021. PMID: 34334388
-
Histiocytic neoplasms in the era of personalized genomic medicine.Curr Opin Hematol. 2016 Jul;23(4):416-25. doi: 10.1097/MOH.0000000000000256. Curr Opin Hematol. 2016. PMID: 27101528 Free PMC article. Review.
Cited by
-
Racial disparities in colorectal cancer clinicopathological and molecular tumor characteristics: a systematic review.Cancer Causes Control. 2024 Feb;35(2):223-239. doi: 10.1007/s10552-023-01783-y. Epub 2023 Sep 9. Cancer Causes Control. 2024. PMID: 37688643 Free PMC article.
-
Incidental germline findings during comprehensive genomic profiling of pancreatic and colorectal cancer: single-centre, molecular tumour board experience.Mutagenesis. 2025 Mar 15;40(1):20-29. doi: 10.1093/mutage/geae014. Mutagenesis. 2025. PMID: 38773787 Free PMC article.
References
-
- NCCN guidelines for colon cancer. [ Mar; 2022 ];https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf 2022
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
