PRI: Re-Analysis of a Public Mass Cytometry Dataset Reveals Patterns of Effective Tumor Treatments
- PMID: 35592315
- PMCID: PMC9110672
- DOI: 10.3389/fimmu.2022.849329
PRI: Re-Analysis of a Public Mass Cytometry Dataset Reveals Patterns of Effective Tumor Treatments
Abstract
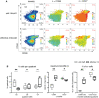
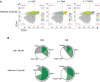
Recently, mass cytometry has enabled quantification of up to 50 parameters for millions of cells per sample. It remains a challenge to analyze such high-dimensional data to exploit the richness of the inherent information, even though many valuable new analysis tools have already been developed. We propose a novel algorithm "pattern recognition of immune cells (PRI)" to tackle these high-dimensional protein combinations in the data. PRI is a tool for the analysis and visualization of cytometry data based on a three or more-parametric binning approach, feature engineering of bin properties of multivariate cell data, and a pseudo-multiparametric visualization. Using a publicly available mass cytometry dataset, we proved that reproducible feature engineering and intuitive understanding of the generated bin plots are helpful hallmarks for re-analysis with PRI. In the CD4+T cell population analyzed, PRI revealed two bin-plot patterns (CD90/CD44/CD86 and CD90/CD44/CD27) and 20 bin plot features for threshold-independent classification of mice concerning ineffective and effective tumor treatment. In addition, PRI mapped cell subsets regarding co-expression of the proliferation marker Ki67 with two major transcription factors and further delineated a specific Th1 cell subset. All these results demonstrate the added insights that can be obtained using the non-cluster-based tool PRI for re-analyses of high-dimensional cytometric data.
Keywords: combinatorial protein expression; high-dimensional cytometry data; mass cytometry data; multi-parametric analysis; pattern perception; re-analysis.
Copyright © 2022 Hoang, Gryzik, Hoppe, Rybak, Schädlich, Kadner, Walther, Vera, Radbruch, Groth, Baumgart and Baumgrass.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

