Exploring Machine Learning Algorithms to Unveil Genomic Regions Associated With Resistance to Southern Root-Knot Nematode in Soybeans
- PMID: 35592556
- PMCID: PMC9111516
- DOI: 10.3389/fpls.2022.883280
Exploring Machine Learning Algorithms to Unveil Genomic Regions Associated With Resistance to Southern Root-Knot Nematode in Soybeans
Abstract
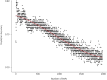
Southern root-knot nematode [SRKN, Meloidogyne incognita (Kofold & White) Chitwood] is a plant-parasitic nematode challenging to control due to its short life cycle, a wide range of hosts, and limited management options, of which genetic resistance is the main option to efficiently control the damage caused by SRKN. To date, a major quantitative trait locus (QTL) mapped on chromosome (Chr.) 10 plays an essential role in resistance to SRKN in soybean varieties. The confidence of discovered trait-loci associations by traditional methods is often limited by the assumptions of individual single nucleotide polymorphisms (SNPs) always acting independently as well as the phenotype following a Gaussian distribution. Therefore, the objective of this study was to conduct machine learning (ML)-based genome-wide association studies (GWAS) utilizing Random Forest (RF) and Support Vector Machine (SVM) algorithms to unveil novel regions of the soybean genome associated with resistance to SRKN. A total of 717 breeding lines derived from 330 unique bi-parental populations were genotyped with the Illumina Infinium BARCSoySNP6K BeadChip and phenotyped for SRKN resistance in a greenhouse. A GWAS pipeline involving a supervised feature dimension reduction based on Variable Importance in Projection (VIP) and SNP detection based on classification accuracy was proposed. Minor effect SNPs were detected by the proposed ML-GWAS methodology but not identified using Bayesian-information and linkage-disequilibrium Iteratively Nested Keyway (BLINK), Fixed and Random Model Circulating Probability Unification (FarmCPU), and Enriched Compressed Mixed Linear Model (ECMLM) models. Besides the genomic region on Chr. 10 that can explain most of SRKN resistance variance, additional minor effects SNPs were also identified on Chrs. 10 and 11. The findings in this study demonstrated that overfitting in GWAS may lead to lower prediction accuracy, and the detection of significant SNPs based on classification accuracy limited false-positive associations. The expansion of the basis of the genetic resistance to SRKN can potentially reduce the selection pressure over the major QTL on Chr. 10 and achieve higher levels of resistance.
Keywords: GWAS; feature selection; machine learning; root-knot nematode; soybean.
Copyright © 2022 Canella Vieira, Zhou, Usovsky, Vuong, Howland, Lee, Li, Zhou, Shannon, Nguyen and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
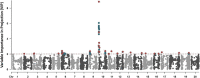
Similar articles
-
Identification of quantitative trait loci underlying resistance to southern root-knot and reniform nematodes in soybean accession PI 567516C.Mol Breed. 2015;35(6):131. doi: 10.1007/s11032-015-0330-5. Epub 2015 May 23. Mol Breed. 2015. PMID: 26028986 Free PMC article.
-
A Quantitative Trait Locus on Maize Chromosome 5 Is Associated with Root-Knot Nematode Resistance.Phytopathology. 2024 Jul;114(7):1657-1663. doi: 10.1094/PHYTO-08-23-0286-R. Epub 2024 Jul 1. Phytopathology. 2024. PMID: 38427606
-
Mapping of Nematode Resistance in Hexaploid Sweetpotato Using an Next-Generation Sequencing-Based Association Study.Front Plant Sci. 2022 Mar 18;13:858747. doi: 10.3389/fpls.2022.858747. eCollection 2022. Front Plant Sci. 2022. PMID: 35371138 Free PMC article.
-
Advancements in breeding, genetics, and genomics for resistance to three nematode species in soybean.Theor Appl Genet. 2016 Dec;129(12):2295-2311. doi: 10.1007/s00122-016-2816-x. Epub 2016 Oct 28. Theor Appl Genet. 2016. PMID: 27796432 Review.
-
Characterization of Disease Resistance Loci in the USDA Soybean Germplasm Collection Using Genome-Wide Association Studies.Phytopathology. 2016 Oct;106(10):1139-1151. doi: 10.1094/PHYTO-01-16-0042-FI. Epub 2016 Jul 11. Phytopathology. 2016. PMID: 27135674 Review.
Cited by
-
Combating Root-Knot Nematodes (Meloidogyne spp.): From Molecular Mechanisms to Resistant Crops.Plants (Basel). 2025 Apr 27;14(9):1321. doi: 10.3390/plants14091321. Plants (Basel). 2025. PMID: 40364350 Free PMC article. Review.
-
Identification of genomic regions associated with soybean responses to off-target dicamba exposure.Front Plant Sci. 2022 Dec 9;13:1090072. doi: 10.3389/fpls.2022.1090072. eCollection 2022. Front Plant Sci. 2022. PMID: 36570921 Free PMC article.
-
Novel genetic resources associated with sucrose and stachyose content through genome-wide association study in soybean (Glycine max (L.) Merr.).Front Plant Sci. 2023 Nov 1;14:1294659. doi: 10.3389/fpls.2023.1294659. eCollection 2023. Front Plant Sci. 2023. PMID: 38023839 Free PMC article.
-
Genetic architecture of soybean tolerance to off-target dicamba.Front Plant Sci. 2023 Oct 9;14:1230068. doi: 10.3389/fpls.2023.1230068. eCollection 2023. Front Plant Sci. 2023. PMID: 37877091 Free PMC article.
References
-
- Akarachantachote N., Chadcham S., Saithanu K. (2014). Cutoff threshold of variable importance in projection for variable selection. Int. J. Pure Appl. Math. 94 307–322. 10.12732/ijpam.v94i3.2 - DOI
-
- Allen T. W., Bradley C. A., Sisson A. J., Byamukama E., Chilvers M. I., Coker C. M., et al. (2017). Soybean yield loss estimates due to diseases in the United States and Ontario, Canada, from 2010 to 2014. Plant Health Prog. 18 19–27. 10.1094/PHP-RS-16-0066 - DOI
-
- Beneventi M. A., da Silva O. B., de Sá M. E. L., Firmino A. A. P., de Amorim R. M. S., Albuquerque ÉV. S., et al. (2013). Transcription profile of soybean-root-knot nematode interaction reveals a key role of phythormones in the resistance reaction. BMC Genomics 14:322. 10.1186/1471-2164-14-322 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

