Cx43-mediated sorting of miRNAs into extracellular vesicles
- PMID: 35593040
- PMCID: PMC9253745
- DOI: 10.15252/embr.202154312
Cx43-mediated sorting of miRNAs into extracellular vesicles
Abstract
Through the exchange of lipids, proteins, and nucleic acids, extracellular vesicles (EV) allow for cell-cell communication across distant cells and tissues to regulate a wide range of physiological and pathological processes. Although some molecular mediators have been discovered, the mechanisms underlying the selective sorting of miRNAs into EV remain elusive. Previous studies demonstrated that connexin43 (Cx43) forms functional channels at the EV surface, mediating the communication with recipient cells. Here, we show that Cx43 participates in the selective sorting of miRNAs into EV through a process that can also involve RNA-binding proteins. We provide evidence that Cx43 can directly bind to specific miRNAs, namely those containing stable secondary structure elements, including miR-133b. Furthermore, Cx43 facilitates the delivery of EV-miRNAs into recipient cells. Phenotypically, we show that Cx43-mediated EV-miRNAs sorting modulates autophagy. Overall, our study ascribes another biological role to Cx43, that is, the selective incorporation of miRNAs into EV, which potentially modulates multiple biological processes in target cells and may have implications for human health and disease.
Keywords: Connexin43; heterogeneous nuclear ribonucleoproteins; intercellular communication; miRNA; selective sorting.
© 2022 The Authors.
Figures

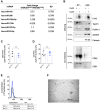
EV produced by HEK293 cells expressing or not Cx43 (EVCx43+ or EVCx43−) were purified by differential ultracentrifugation. miRNAs expression profile analysis in EVCx43+ (represented as fold increase over EVCx43−).
Pathway enrichment analysis for the predicted targets of the over‐represented miRNAs in Cx43‐containing EV.
HEK293 cells were incubated with 5 µg of purified EVCx43− or EVCx43+ for 24 h, in the presence or absence of Bafilomycin A1 (Baf) in the last 6 h. The levels of LC3 were analyzed by WB and are depicted on graph (LC3‐II; n = 4 biological replicates).
miRNA‐centered regulatory network established by the miRNAs enriched in Cx43‐containing EV. The figure represents predicted targets that are simultaneously regulated by 3 or more of the selected miRNAs. The size of the symbols is proportional to the number of established functional connections. miRNA‐regulatory are represented by grey edges. Protein–protein interactions obtained from STRING database are depicted in black lines.
HEK293 cells were incubated with EVCx43− or EVCx43+ for 24 h before WB analysis of ATP2A2/SERCA2, a predicted target of miRNAs enriched in Cx43‐containing EV. Graph depicts quantification of ATP2A2/SERCA2 levels (n = 4 biological replicates).
miRNAs expression profile analysis in HEK293Cx43+ cells (represented as fold increase over HEK293Cx43− cells).
EV produced by HEK293 cells expressing or not Cx43 (EVCx43+ or EVCx43−) were purified by differential ultracentrifugation. WB analysis of positive EV markers (Tsg101, Flotillin‐1 and GAPDH) and the negative marker Calnexin. WB for CD63 and CD81 was performed in non‐reducing conditions. Cellular extracts were used as control. 10 μg of total protein were loaded in each case.
Graph depicts total protein levels in purified EV produced by HEK293 cells expressing or not Cx43 (EVCx43+ or EVCx43−; n = 3 biological replicates).
Graph depicts total RNA levels in purified EV produced by HEK293 cells expressing or not Cx43 (EVCx43+ or EVCx43−; n = 4 biological replicates).
Representative NTA of concentration and size distribution of EV produced by HEK293 cells expressing or not Cx43 (EVCx43+ and EVCx43−; n = 2 biological replicates).
Representative TEM image of purified EVCx43+.
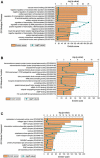
GO Biological Process.
GO Molecular Function.
GO Cellular Component.
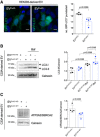
HEK293 cells transfected with GFP‐LC3 were incubated with EVCx43− or EVCx43+ for 24 h. In the last 6 h, cells were serum‐starved before analysis of the number of GFP‐LC3 puncta. Scale bars 10 μm. Graph depicts the average number of GFP‐LC3 puncta per analyzed cell (n = 4 biological replicates).
Parental C33a cells were incubated with 5 µg of purified EV obtained from parental (EVCx43‐WT) or Cx43‐knockout cells (EVCx43‐KO) for 24 h, in the presence or absence of Bafilomycin A1 in the last 6 h. The levels of LC3 were analyzed by WB and depicted on graph (LC3‐II; n = 4 biological replicates).
Parental C33a cells were incubated with EVCx43‐WT or EVCx43‐KO for 24 h before WB analysis of ATP2A2/SERCA2, a predicted target of miRNAs enriched in Cx43‐containing EV. Quantification of the levels of ATP2A2/SERCA2 is depicted on graph (n = 4 biological replicates).

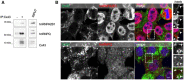
Cx43 was immunoprecipitated (IP) from HEK293Cx43+ cells, after which interaction with hnRNPA2B1 and hnRNPQ was analyzed by WB.
Co‐localization between Cx43 (green) and hnRNPA2B1 or hnRNPQ (red) in C33a cells was analyzed by confocal microscopy. Scale bars 10 μm. Arrowheads in the inset images highlight co‐localization between Cx43 and hnRNPs.
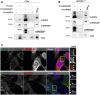
IP of Cx43, hnRNPA2B1, or hnRNPQ was performed in C33a cells, as indicated. Interaction with hnRNPA2B1, hnRNPQ, and Cx43 was analyzed by WB.
hnRNPA2B1 or hnRNPQ were IP from HEK293Cx43+ cells, where indicated, after which interaction with Cx43 was analyzed by WB.
Co‐localization between Cx43 (green) and hnRNPA2B1 or hnRNPQ (red) in HEK293Cx43+ cells was analyzed by confocal microscopy. Scale bars 10 μm. Arrowheads in the inset images highlight co‐localization between Cx43 and hnRNPs.
- A–D
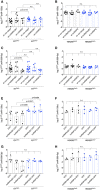
EV‐producing cells expressing or not Cx43 (HEK293Cx43+ or HEK293Cx43−) were transiently transfected with shRNAs targeting hnRNPA2B1, hnRNPQ or non‐target shRNAs, for 48 h. miR‐133b levels were assessed in EVCx43+ and EVCx43− (A; n = 6–12 biological replicates) and EV‐producing cells (B; n = 4–9 biological replicates). miR‐509‐3p levels were assessed in EVCx43+ and EVCx43− (C; n = 6–15 biological replicates) and EV‐producing cells (D; n = 4–9 biological replicates).
- E–H
HEK293Cx43+ or HEK293Cx43− were transiently transfected with hnRNPA2B1‐GFP, GFP‐hnRNPQ or GFP alone, for 24 h. miR‐133b levels were assessed in EVCx43+ and EVCx43− (E; n = 4 biological replicates) and EV‐producing cells (F; n = 3 biological replicates). miR‐509‐3p levels were assessed in EVCx43+ and EVCx43− (G; n = 4 biological replicates) and EV‐producing cells (H; n = 3 biological replicates).
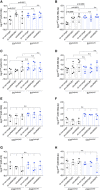
- A–H
EV‐producing C33a cells expressing or not Cx43 (C33a‐WT or C33a‐KO) were transiently transfected with shRNAs targeting hnRNPA2B1, hnRNPQ or non‐target shRNAs, for 48 h. Levels of miR‐133b (A), miR‐199‐3p (B), miR‐410 (C) and miR‐509‐3p (D) were assessed in EVCx43‐WT and EVCx43‐KO (n = 4–5 biological replicates). Levels of miR‐133b (E), miR‐199‐3p (F), miR‐410 (G) and miR‐509‐3p (H) were assessed in C33a cells (n = 3 biological replicates).

- A
Cell free EV biogenesis was assessed by relative protection of Cx43‐luciferase. Reactions were incubated at 4 or 30°C in the presence of membranes derived from HEK293Cx43− transiently transfected with Cx43‐luciferase and cytosol from HEK293Cx43− cells, except in the negative control. 1% TX100 was used to disrupt membranes, where indicated (n = 3 biological replicates).
- B
Crude membranes and cytosol from broken HEK293Cx43+ or HEK293Cx43− cells were mixed in an ATP regenerating system, supplemented with synthetic miR‐133b or miR‐509‐3p, as indicated. Graph depicts the indicated miRNAs levels, assessed by RT‐qPCR and represented as percent protected after RNAse I digestion of unincorporated miRNAs (n = 7–8 biological replicates).
- C
Co‐immunoprecipitation of endogenous miR‐133b and miR‐509‐3p following UV crosslinking immunoprecipitation (CLIP) of Cx43 (n = 3–5 biological replicates).
- D
WB analysis for Cx43 in samples derived by miRNAs pulldowns performed with whole lysates of HEK293Cx43+ cells and biotinylated miR‐133b and miR‐509‐3p (n = 5 biological replicates).
- E–H
EMSA was performed using either Cy3‐labeled miR‐133b (E,F) or miR‐509‐3p (G,H), and Cx43 produced by in vitro translation inserted into lipid nanodiscs, in the presence or absence of recombinant GST‐hnRNPA2B1 or GST‐hnRNPQ, as indicated. For competition EMSA, an excess of unlabeled miR‐133b or miR‐509‐3p was used (competitor). Empty nanodiscs were used to control unspecific binding.
- I
EMSA was performed using either Cy3‐labeled wild‐type or mutant miR‐199a‐3p, and Cx43‐enriched membranes prepared from C33a parental cells (Cx43+). Cx43− membranes were used to control unspecific binding.
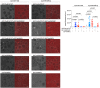
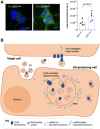
Equal amounts of EVCx43+ and EVCx43− labelled with SYTO® RNASelect™ green fluorescent stain were incubated with HEK293Cx43+ cells for 30 min, at 37°C. Scale bars 10 μm (n = 5 biological replicates). P‐values were derived by Mann‐Whitney test.
Working model for the role of Cx43 in mediating the selective sorting of miRNAs into EV. Cx43 preferentially binds to miRNAs harboring a stable secondary structure, mediating its selective incorporation into EV. Although hnRNPs do not seem to directly participate in Cx43‐dependent loading of EV‐miRNAs, they strength the binding of miRNAs to cx43, suggesting that a multiprotein complex formed between RNA‐binding proteins and Cx43 is important to select miRNAs.
References
-
- Baer C, Lyle R, Burdet F, Gilfillan GD, De Palma M, Squadrito ML, Ibberson M, Maderna C (2014) Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep 8: 1432–1446 - PubMed
-
- Catarino S, Ramalho JS, Marques C, Pereira P, Girão H (2011) Ubiquitin‐mediated internalization of connexin43 is independent of the canonical endocytic tyrosine‐sorting signal. Biochem J 437: 255–267 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

