Snrpb is required in murine neural crest cells for proper splicing and craniofacial morphogenesis
- PMID: 35593225
- PMCID: PMC9235875
- DOI: 10.1242/dmm.049544
Snrpb is required in murine neural crest cells for proper splicing and craniofacial morphogenesis
Abstract
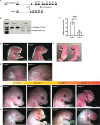
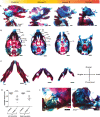
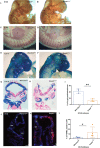
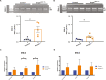
Heterozygous mutations in SNRPB, an essential core component of the five small ribonucleoprotein particles of the spliceosome, are responsible for cerebrocostomandibular syndrome (CCMS). We show that Snrpb heterozygous mouse embryos arrest shortly after implantation. Additionally, heterozygous deletion of Snrpb in the developing brain and neural crest cells models craniofacial malformations found in CCMS, and results in death shortly after birth. RNAseq analysis of mutant heads prior to morphological defects revealed increased exon skipping and intron retention in association with increased 5' splice site strength. We found increased exon skipping in negative regulators of the P53 pathway, along with increased levels of nuclear P53 and P53 target genes. However, removing Trp53 in Snrpb heterozygous mutant neural crest cells did not completely rescue craniofacial development. We also found a small but significant increase in exon skipping of several transcripts required for head and midface development, including Smad2 and Rere. Furthermore, mutant embryos exhibited ectopic or missing expression of Fgf8 and Shh, which are required to coordinate face and brain development. Thus, we propose that mis-splicing of transcripts that regulate P53 activity and craniofacial-specific genes contributes to craniofacial malformations. This article has an associated First Person interview with the first author of the paper.
Keywords: SNRPB; Cerebrocostomandibular syndrome; Craniofacial; Neural crest cells; Splicing.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures



References
-
- Bacrot, S., Doyard, M., Huber, C., Alibeu, O., Feldhahn, N., Lehalle, D., Lacombe, D., Marlin, S., Nitschke, P., Petit, F.et al. (2015). Mutations in SNRPB, encoding components of the core splicing machinery, cause cerebro-costo-mandibular syndrome. Hum. Mutat. 36, 187-190. 10.1002/humu.22729 - DOI - PubMed
-
- Beauchamp, M.-C., Djedid, A., Bareke, E., Merkuri, F., Aber, R., Tam, A. S., Lines, M. A., Boycott, K. M., Stirling, P. C., Fish, J. L.et al. (2021). Mutation in Eftud2 causes craniofacial defects in mice via mis-splicing of Mdm2 and increased P53. Hum. Mol. Genet. 30, 739-757. 10.1093/hmg/ddab051 - DOI - PMC - PubMed
-
- Bogue, M. A., Grubb, S. C., Walton, D. O., Philip, V. M., Kolishovski, G., Stearns, T., Dunn, M. H., Skelly, D. A., Kadakkuzha, B., TeHennepe, G.et al. (2018). Mouse Phenome Database: an integrative database and analysis suite for curated empirical phenotype data from laboratory mice. Nucleic Acids Res. 46, D843-D850. 10.1093/nar/gkx1082 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

