Exploring the onset of B12 -based mutualisms using a recently evolved Chlamydomonas auxotroph and B12 -producing bacteria
- PMID: 35593514
- PMCID: PMC9545926
- DOI: 10.1111/1462-2920.16035
Exploring the onset of B12 -based mutualisms using a recently evolved Chlamydomonas auxotroph and B12 -producing bacteria
Erratum in
-
Correction to "Exploring the onset of B12-based mutualisms using a recently evolved Chlamydomonas auxotroph and B12-producing bacteria".Environ Microbiol. 2024 Feb;26(2):e16575. doi: 10.1111/1462-2920.16575. Epub 2024 Jan 19. Environ Microbiol. 2024. PMID: 38240321 Free PMC article. No abstract available.
Abstract
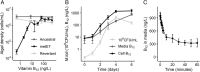
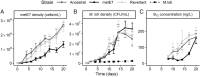
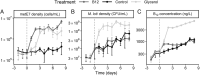
Cobalamin (vitamin B12 ) is a cofactor for essential metabolic reactions in multiple eukaryotic taxa, including major primary producers such as algae, and yet only prokaryotes can produce it. Many bacteria can colonize the algal phycosphere, forming stable communities that gain preferential access to photosynthate and in return provide compounds such as B12 . Extended coexistence can then drive gene loss, leading to greater algal-bacterial interdependence. In this study, we investigate how a recently evolved B12 -dependent strain of Chlamydomonas reinhardtii, metE7, forms a mutualism with certain bacteria, including the rhizobium Mesorhizobium loti and even a strain of the gut bacterium E. coli engineered to produce cobalamin. Although metE7 was supported by B12 producers, its growth in co-culture was slower than the B12 -independent wild-type, suggesting that high bacterial B12 provision may be necessary to favour B12 auxotrophs and their evolution. Moreover, we found that an E. coli strain that releases more B12 makes a better mutualistic partner, and although this trait may be more costly in isolation, greater B12 release provided an advantage in co-cultures. We hypothesize that, given the right conditions, bacteria that release more B12 may be selected for, particularly if they form close interactions with B12 -dependent algae.
© 2022 The Authors. Environmental Microbiology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Figures


References
-
- Acevedo‐Rocha, C.G. , Gronenberg, L.S. , Mack, M. , Commichau, F.M. , and Genee, H.J. (2019) Microbial cell factories for the sustainable manufacturing of B vitamins. Curr Opin Biotechnol 56: 18–29. - PubMed
-
- Amin, S.A. , Hmelo, L.R. , van Tol, H.M. , Durham, B.P. , Carlson, L.T. , Heal, K.R. , et al. (2015) Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522: 98–101. - PubMed
-
- Barber‐Lluch, E. , Joglar, V. , Moreiras, G. , Leão, J.M. , Gago‐Martínez, A. , Fernández, E. , and Teira, E. (2021) Variability of vitamin B12 concentrations in waters along the northwest Iberian shelf. Reg Stud Mar Sci 42: 101608.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- BB/D011043/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I013164/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/S002197/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M011194/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

