The long noncoding RNA Malat1 regulates CD8+ T cell differentiation by mediating epigenetic repression
- PMID: 35593887
- PMCID: PMC9127983
- DOI: 10.1084/jem.20211756
The long noncoding RNA Malat1 regulates CD8+ T cell differentiation by mediating epigenetic repression
Abstract
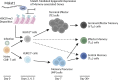
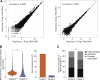
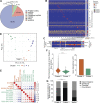
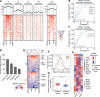
During an immune response to microbial infection, CD8+ T cells give rise to short-lived effector cells and memory cells that provide sustained protection. Although the transcriptional programs regulating CD8+ T cell differentiation have been extensively characterized, the role of long noncoding RNAs (lncRNAs) in this process remains poorly understood. Using a functional genetic knockdown screen, we identified the lncRNA Malat1 as a regulator of terminal effector cells and the terminal effector memory (t-TEM) circulating memory subset. Evaluation of chromatin-enriched lncRNAs revealed that Malat1 grouped with trans lncRNAs that exhibit increased RNA interactions at gene promoters and gene bodies. Moreover, we observed that Malat1 was associated with increased H3K27me3 deposition at a number of memory cell-associated genes through a direct interaction with Ezh2, thereby promoting terminal effector and t-TEM cell differentiation. Our findings suggest an important functional role of Malat1 in regulating CD8+ T cell differentiation and broaden the knowledge base of lncRNAs in CD8+ T cell biology.
© 2022 Kanbar et al.
Conflict of interest statement
Disclosures: G.W. Yeo is a co-founder, member of the Board of Directors, on the SAB, equity holder, and paid consultant for Locanabio and Eclipse BioInnovations. In addition, G.W. Yeo is a visiting professor at the National University of Singapore. G.W. Yeo’s interest(s) have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. J.T. Chang reported grants from Takeda and Eli Lilly outside the submitted work. No other disclosures were reported.
Figures







References
-
- Banerjee, A., Gordon S.M., Intlekofer A.M., Paley M.A., Mooney E.C., Lindsten T., Wherry E.J., and Reiner S.L.. 2010. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J. Immunol. 185:4988–4992. 10.4049/jimmunol.1002042 - DOI - PMC - PubMed
-
- Boland, B.S., He Z., Tsai M.S., Olvera J.G., Omilusik K.D., Duong H.G., Kim E.S., Limary A.E., Jin W., Milner J.J., et al. 2020. Heterogeneity and clonal relationships of adaptive immune cells in ulcerative colitis revealed by single-cell analyses. Sci Immunol. 5:eabb4432. 10.1126/sciimmunol.abb4432 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R56 DK098808/DK/NIDDK NIH HHS/United States
- R01 DK098808/DK/NIDDK NIH HHS/United States
- R01 AI129973/AI/NIAID NIH HHS/United States
- P30 DK120515/DK/NIDDK NIH HHS/United States
- S10 OD026929/OD/NIH HHS/United States
- R01 HG004659/HG/NHGRI NIH HHS/United States
- R01 AI123202/AI/NIAID NIH HHS/United States
- P30KC063491/NH/NIH HHS/United States
- I01 BX005106/BX/BLRD VA/United States
- P30 NS047101/NS/NINDS NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- R01 GM124494/GM/NIGMS NIH HHS/United States
- P30DK120515/San Diego Digestive Diseases Research Center
- P01 AI132122/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

