MERS-CoV endoribonuclease and accessory proteins jointly evade host innate immunity during infection of lung and nasal epithelial cells
- PMID: 35594398
- PMCID: PMC9173776
- DOI: 10.1073/pnas.2123208119
MERS-CoV endoribonuclease and accessory proteins jointly evade host innate immunity during infection of lung and nasal epithelial cells
Abstract
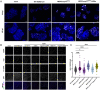
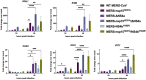
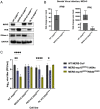
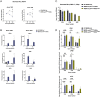
Middle East respiratory syndrome coronavirus (MERS-CoV) emerged into humans in 2012, causing highly lethal respiratory disease. The severity of disease may be, in part, because MERS-CoV is adept at antagonizing early innate immune pathways—interferon (IFN) production and signaling, protein kinase R (PKR), and oligoadenylate synthetase/ribonuclease L (OAS/RNase L)—activated in response to viral double-stranded RNA (dsRNA) generated during genome replication. This is in contrast to severe acute respiratory syndrome CoV-2 (SARS-CoV-2), which we recently reported to activate PKR and RNase L and, to some extent, IFN signaling. We previously found that MERS-CoV accessory proteins NS4a (dsRNA binding protein) and NS4b (phosphodiesterase) could weakly suppress these pathways, but ablation of each had minimal effect on virus replication. Here we investigated the antagonist effects of the conserved coronavirus endoribonuclease (EndoU), in combination with NS4a or NS4b. Inactivation of EndoU catalytic activity alone in a recombinant MERS-CoV caused little if any effect on activation of the innate immune pathways during infection. However, infection with recombinant viruses containing combined mutations with inactivation of EndoU and deletion of NS4a or inactivation of the NS4b phosphodiesterase promoted robust activation of dsRNA-induced innate immune pathways. This resulted in at least tenfold attenuation of replication in human lung–derived A549 and primary nasal cells. Furthermore, replication of these recombinant viruses could be rescued to the level of wild-type MERS-CoV by knockout of host immune mediators MAVS, PKR, or RNase L. Thus, EndoU and accessory proteins NS4a and NS4b together suppress dsRNA-induced innate immunity during MERS-CoV infection in order to optimize viral replication.
Keywords: MERS-CoV; PKR; RNase L; endonuclease U; innate immune antagonism.
Conflict of interest statement
Competing interest statement: S.R.W. is on the Scientific Advisory Boards of Immunome, Inc and Ocugen, Inc. N.A.C. consults for GSK, AstraZeneca, Novartis, Sanofi/Regeneron, and Oyster Point Pharmaceuticals and has US patent “Therapy and Diagnostics for Respiratory Infection” (10,881,698 B2, WO20913112865) and a licensing agreement with GeneOne Life Sciences. A.R.F. consults with Deciphera Inc.
Figures


Update of
-
MERS-CoV endoribonuclease and accessory proteins jointly evade host innate immunity during infection of lung and nasal epithelial cells.bioRxiv [Preprint]. 2021 Dec 21:2021.12.20.473564. doi: 10.1101/2021.12.20.473564. bioRxiv. 2021. Update in: Proc Natl Acad Sci U S A. 2022 May 24;119(21):e2123208119. doi: 10.1073/pnas.2123208119. PMID: 34981054 Free PMC article. Updated. Preprint.
References
-
- Zaki A. M., van Boheemen S., Bestebroer T. M., Osterhaus A. D., Fouchier R. A., Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

