R-BIND 2.0: An Updated Database of Bioactive RNA-Targeting Small Molecules and Associated RNA Secondary Structures
- PMID: 35594415
- PMCID: PMC9343015
- DOI: 10.1021/acschembio.2c00224
R-BIND 2.0: An Updated Database of Bioactive RNA-Targeting Small Molecules and Associated RNA Secondary Structures
Abstract
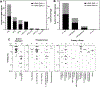
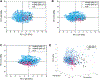
Discoveries of RNA roles in cellular physiology and pathology are increasing the need for new tools that modulate the structure and function of these biomolecules, and small molecules are proving useful. In 2017, we curated the
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

