Gut dysbiosis in rheumatic diseases: A systematic review and meta-analysis of 92 observational studies
- PMID: 35594658
- PMCID: PMC9120231
- DOI: 10.1016/j.ebiom.2022.104055
Gut dysbiosis in rheumatic diseases: A systematic review and meta-analysis of 92 observational studies
Abstract
Background: Emerging evidence suggests that dysbiosis in gut microbiota may contribute to the occurrence or development of several rheumatic diseases. Since gut microbiota dysbiosis is potentially modifiable, it has been postulated to be a promising preventive or therapeutic target for rheumatic diseases. However, the current understanding on the potential associations between gut microbiota and rheumatic diseases is still inadequate. Therefore, we aimed to synthesise the accumulating evidence for the relation of gut microbiota to rheumatic diseases.
Methods: The PubMed, Embase and Cochrane Library were searched from inception to March 11, 2022 to include observational studies evaluating the associations between gut microbiota and rheumatic diseases. Standardised mean difference (SMD) of α-diversity indices between rheumatic diseases and controls were estimated using random-effects model. β-diversity indices and relative abundance of gut microbes were summarised qualitatively.
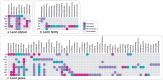
Findings: Of the included 92 studies (11,998 participants), 68 provided data for α-diversity. Taken together as a whole, decreases in α-diversity indices were consistently found in rheumatic diseases (observed species: SMD = -0.36, [95%CI = -0.63, -0.09]; Chao1: SMD = -0.57, [95%CI = -0.88, -0.26]; Shannon index: SMD = -0.33, [95%CI = -0.48, -0.17]; Simpson index: SMD = -0.32, [95%CI = -0.49, -0.14]). However, when specific rheumatic diseases were examined, decreases were only observed in rheumatoid arthritis (observed species: SMD = -0.51, [95%CI = -0.78, -0.24]; Shannon index: SMD = -0.31, [95%CI = -0.49, -0.13]; Simpson index: SMD = -0.31, [95%CI = -0.54, -0.08]), systemic lupus erythematosus (Chao1: SMD = -1.60, [95%CI = -2.54, -0.66]; Shannon index: SMD = -0.63, [95%CI = -1.08, -0.18]), gout (Simpson index: SMD = -0.64, [95%CI = -1.07, -0.22]) and fibromyalgia (Simpson index: SMD = -0.28, [95%CI = -0.44, -0.11]), whereas an increase was observed in systemic sclerosis (Shannon index: SMD = 1.25, [95%CI = 0.09, 2.41]). Differences with statistical significance in β-diversity were consistently reported in ankylosing spondylitis and IgG4-related diseases. Although little evidence of disease specificity of gut microbes was found, shared alterations of the depletion of anti-inflammatory butyrate-producing microbe (i.e., Faecalibacterium) and the enrichment of pro-inflammatory microbe (i.e., Streptococcus) were observed in rheumatoid arthritis, Sjögren's syndrome and systemic lupus erythematosus.
Interpretation: Gut microbiota dysbiosis was associated with rheumatic diseases, principally with potentially non-specific, shared alterations of microbes.
Funding: National Natural Science Foundation of China (81930071, 81902265, 82072502 and U21A20352).
Keywords: Gut dysbiosis; Gut microbiota; Meta-analysis; Rheumatic diseases.
Copyright © 2022 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
No conflict of interest for any of the authors.
Figures



References
-
- Kamada N., Seo S.U., Chen G.Y., Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13(5):321–335. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

