Human NLRP1 is a sensor of pathogenic coronavirus 3CL proteases in lung epithelial cells
- PMID: 35594856
- PMCID: PMC9108100
- DOI: 10.1016/j.molcel.2022.04.033
Human NLRP1 is a sensor of pathogenic coronavirus 3CL proteases in lung epithelial cells
Abstract
Inflammation observed in SARS-CoV-2-infected patients suggests that inflammasomes, proinflammatory intracellular complexes, regulate various steps of infection. Lung epithelial cells express inflammasome-forming sensors and constitute the primary entry door of SARS-CoV-2. Here, we describe that the NLRP1 inflammasome detects SARS-CoV-2 infection in human lung epithelial cells. Specifically, human NLRP1 is cleaved at the Q333 site by multiple coronavirus 3CL proteases, which triggers inflammasome assembly and cell death and limits the production of infectious viral particles. Analysis of NLRP1-associated pathways unveils that 3CL proteases also inactivate the pyroptosis executioner Gasdermin D (GSDMD). Subsequently, caspase-3 and GSDME promote alternative cell pyroptosis. Finally, analysis of pyroptosis markers in plasma from COVID-19 patients with characterized severe pneumonia due to autoantibodies against, or inborn errors of, type I interferons (IFNs) highlights GSDME/caspase-3 as potential markers of disease severity. Overall, our findings identify NLRP1 as a sensor of SARS-CoV-2 infection in lung epithelia.
Keywords: 3CL proteases; Gasdermins; NLRP1 inflammasome; SARS-CoV-2; epithelial cells; pyroptosis.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
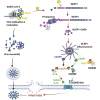
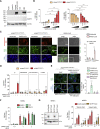


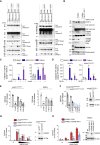
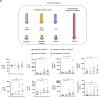
References
-
- Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science. 2003;300:1763–1767. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

