Multiancestral polygenic risk score for pediatric asthma
- PMID: 35595084
- PMCID: PMC9643615
- DOI: 10.1016/j.jaci.2022.03.035
Multiancestral polygenic risk score for pediatric asthma
Abstract
Background: Asthma is the most common chronic condition in children and the third leading cause of hospitalization in pediatrics. The genome-wide association study catalog reports 140 studies with genome-wide significance. A polygenic risk score (PRS) with predictive value across ancestries has not been evaluated for this important trait.
Objectives: This study aimed to train and validate a PRS relying on genetic determinants for asthma to provide predictions for disease occurrence in pediatric cohorts of diverse ancestries.
Methods: This study applied a Bayesian regression framework method using the Trans-National Asthma Genetic Consortium genome-wide association study summary statistics to derive a multiancestral PRS score, used one Electronic Medical Records and Genomics (eMERGE) cohort as a training set, used a second independent eMERGE cohort to validate the score, and used the UK Biobank data to replicate the findings. A phenome-wide association study was performed using the PRS to identify shared genetic etiology with other phenotypes.
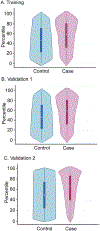
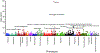
Results: The multiancestral asthma PRS was associated with asthma in the 2 pediatric validation datasets. Overall, the multiancestral asthma PRS has an area under the curve (AUC) of 0.70 (95% CI, 0.69-0.72) in the pediatric validation 1 and AUC of 0.66 (0.65-0.66) in the pediatric validation 2 datasets. We found significant discrimination across pediatric subcohorts of European (AUC, 95% CI, 0.60 and 0.66), African (AUC, 95% CI, 0.61 and 0.66), admixed American (AUC, 0.64 and 0.70), Southeast Asian (AUC, 0.65), and East Asian (AUC, 0.73) ancestry. Pediatric participants with the top 5% PRS had 2.80 to 5.82 increased odds of asthma compared to the bottom 5% across the training, validation 1, and validation 2 cohorts when adjusted for ancestry. Phenome-wide association study analysis confirmed the strong association of the identified PRS with asthma (odds ratio, 2.71, PFDR = 3.71 × 10-65) and related phenotypes.
Conclusions: A multiancestral PRS for asthma based on Bayesian posterior genomic effect sizes identifies increased odds of pediatric asthma.
Keywords: GWAS; Genetics; PRS; PheWAS; asthma; polygenic risk score.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Competing interests:
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Ramsahai JM, Hansbro PM, Wark PAB. Mechanisms and Management of Asthma Exacerbations. Am J Respir Crit Care Med. 2019;199(4):423–32. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 NS099068/NS/NINDS NIH HHS/United States
- R01 HL132153/HL/NHLBI NIH HHS/United States
- U01 HG006382/HG/NHGRI NIH HHS/United States
- R01 DK107502/DK/NIDDK NIH HHS/United States
- R01 AI139126/AI/NIAID NIH HHS/United States
- R01 HG010730/HG/NHGRI NIH HHS/United States
- U01 HG008679/HG/NHGRI NIH HHS/United States
- UG3 OD023282/OD/NIH HHS/United States
- P30 AR070549/AR/NIAMS NIH HHS/United States
- R01 AI148276/AI/NIAID NIH HHS/United States
- U01 HG006385/HG/NHGRI NIH HHS/United States
- R21 HL145422/HL/NHLBI NIH HHS/United States
- U01 HG008676/HG/NHGRI NIH HHS/United States
- R01 HL132344/HL/NHLBI NIH HHS/United States
- U01 HG006375/HG/NHGRI NIH HHS/United States
- U01 HG004438/HG/NHGRI NIH HHS/United States
- U01 HG011167/HG/NHGRI NIH HHS/United States
- U01 AI130830/AI/NIAID NIH HHS/United States
- R01 AR073228/AR/NIAMS NIH HHS/United States
- U01 HG011172/HG/NHGRI NIH HHS/United States
- R01 AI141569/AI/NIAID NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- U01 HG008657/HG/NHGRI NIH HHS/United States
- U01 AI150748/AI/NIAID NIH HHS/United States
- R01 NS096053/NS/NINDS NIH HHS/United States
- U01 HG004603/HG/NHGRI NIH HHS/United States
- R01 HD089458/HD/NICHD NIH HHS/United States
- R01 HG010166/HG/NHGRI NIH HHS/United States
- U01 HG004424/HG/NHGRI NIH HHS/United States
- U01 HG004609/HG/NHGRI NIH HHS/United States
- U01 HG006389/HG/NHGRI NIH HHS/United States
- R25 GM129808/GM/NIGMS NIH HHS/United States
- U01 HG011175/HG/NHGRI NIH HHS/United States
- U01 HG008672/HG/NHGRI NIH HHS/United States
- U54 AI117804/AI/NIAID NIH HHS/United States
- U01 HG004599/HG/NHGRI NIH HHS/United States
- R01 HD099775/HD/NICHD NIH HHS/United States
- U01 HG008684/HG/NHGRI NIH HHS/United States
- U01 HG011169/HG/NHGRI NIH HHS/United States
- U01 HG006828/HG/NHGRI NIH HHS/United States
- R01 GM055479/GM/NIGMS NIH HHS/United States
- U01 HG006380/HG/NHGRI NIH HHS/United States
- R01 AI024717/AI/NIAID NIH HHS/United States
- U01 HG008666/HG/NHGRI NIH HHS/United States
- U01 HG006388/HG/NHGRI NIH HHS/United States
- U01 HG008680/HG/NHGRI NIH HHS/United States
- U19 AI070235/AI/NIAID NIH HHS/United States
- U01 HG011176/HG/NHGRI NIH HHS/United States
- U01 HG006378/HG/NHGRI NIH HHS/United States
- U01 HG004610/HG/NHGRI NIH HHS/United States
- U01 HG008673/HG/NHGRI NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- R01 HG011411/HG/NHGRI NIH HHS/United States
- U01 HG008685/HG/NHGRI NIH HHS/United States
- U01 HG006379/HG/NHGRI NIH HHS/United States
- U01 HG011166/HG/NHGRI NIH HHS/United States
- U01 HG008664/HG/NHGRI NIH HHS/United States
- R01 HL135114/HL/NHLBI NIH HHS/United States
- U01 HG011181/HG/NHGRI NIH HHS/United States
- R01 AI127392/AI/NIAID NIH HHS/United States
- U01 HG004608/HG/NHGRI NIH HHS/United States
- U01 HG008701/HG/NHGRI NIH HHS/United States
- U01 HG006830/HG/NHGRI NIH HHS/United States

