Validation of an Allosteric Binding Site of Src Kinase Identified by Unbiased Ligand Binding Simulations
- PMID: 35595169
- PMCID: PMC9713948
- DOI: 10.1016/j.jmb.2022.167628
Validation of an Allosteric Binding Site of Src Kinase Identified by Unbiased Ligand Binding Simulations
Abstract
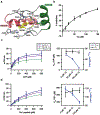
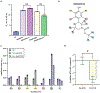
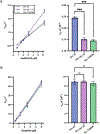
Allostery plays a primary role in regulating protein activity, making it an important mechanism in human disease and drug discovery. Identifying allosteric regulatory sites to explore their biological significance and therapeutic potential is invaluable to drug discovery; however, identification remains a challenge. Allosteric sites are often "cryptic" without clear geometric or chemical features. Since allosteric regulatory sites are often less conserved in protein kinases than the orthosteric ATP binding site, allosteric ligands are commonly more specific than ATP competitive inhibitors. We present a generalizable computational protocol to predict allosteric ligand binding sites based on unbiased ligand binding simulation trajectories. We demonstrate the feasibility of this protocol by revisiting our previously published ligand binding simulations using the first identified viral proto-oncogene, Src kinase, as a model system. The binding paths for kinase inhibitor PP1 uncovered three metastable intermediate states before binding the high-affinity ATP-binding pocket, revealing two previously known allosteric sites and one novel site. Herein, we validate the novel site using a combination of virtual screening and experimental assays to identify a V-type allosteric small-molecule inhibitor that targets this novel site with specificity for Src over closely related kinases. This study provides a proof-of-concept for employing unbiased ligand binding simulations to identify cryptic allosteric binding sites and is widely applicable to other protein-ligand systems.
Keywords: NMR; cancer; docking; drug binding process; inhibitor.
Published by Elsevier Ltd.
Conflict of interest statement
Declaration of interests The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

