A mechanistic study of the influence of nitrogen and energy availability on the NH4+ sensitivity of nitrogen assimilation in Synechococcus
- PMID: 35595516
- PMCID: PMC9467657
- DOI: 10.1093/jxb/erac219
A mechanistic study of the influence of nitrogen and energy availability on the NH4+ sensitivity of nitrogen assimilation in Synechococcus
Abstract
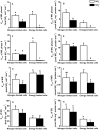
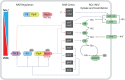
In most algae, NO3- assimilation is tightly controlled and is often inhibited by the presence of NH4+. In the marine, non-colonial, non-diazotrophic cyanobacterium Synechococcus UTEX 2380, NO3- assimilation is sensitive to NH4+ only when N does not limit growth. We sequenced the genome of Synechococcus UTEX 2380, studied the genetic organization of the nitrate assimilation related (NAR) genes, and investigated expression and kinetics of the main NAR enzymes, under N or light limitation. We found that Synechococcus UTEX 2380 is a β-cyanobacterium with a full complement of N uptake and assimilation genes and NAR regulatory elements. The nitrate reductase of our strain showed biphasic kinetics, previously observed only in freshwater or soil diazotrophic Synechococcus strains. Nitrite reductase and glutamine synthetase showed little response to our growth treatments, and their activity was usually much higher than that of nitrate reductase. NH4+ insensitivity of NAR genes may be associated with the stimulation of the binding of the regulator NtcA to NAR gene promoters by the high 2-oxoglutarate concentrations produced under N limitation. NH4+ sensitivity in energy-limited cells fits with the fact that, under these conditions, the use of NH4+ rather than NO3- decreases N-assimilation cost, whereas it would exacerbate N shortage under N limitation.
Keywords: Ammonium; N metabolism; NtcA regulation; cyanobacteria; glutamine synthetase; limitation; nitrate reductase; nitrite reductase.
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society for Experimental Biology. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures





Similar articles
-
Nitrate/nitrite assimilation system of the marine picoplanktonic cyanobacterium Synechococcus sp. strain WH 8103: effect of nitrogen source and availability on gene expression.Appl Environ Microbiol. 2003 Dec;69(12):7009-18. doi: 10.1128/AEM.69.12.7009-7018.2003. Appl Environ Microbiol. 2003. PMID: 14660343 Free PMC article.
-
Role of the Synechococcus PCC 7942 nitrogen regulator protein PipX in NtcA-controlled processes.Microbiology (Reading). 2007 Mar;153(Pt 3):711-718. doi: 10.1099/mic.0.2006/003574-0. Microbiology (Reading). 2007. PMID: 17322191
-
Lack of control of nitrite assimilation by ammonium in an oceanic picocyanobacterium, Synechococcus sp. strain WH 8103.Appl Environ Microbiol. 2007 May;73(9):3028-33. doi: 10.1128/AEM.02606-06. Epub 2007 Mar 2. Appl Environ Microbiol. 2007. PMID: 17337543 Free PMC article.
-
Regulation of nitrate assimilation in cyanobacteria.J Exp Bot. 2011 Feb;62(4):1411-24. doi: 10.1093/jxb/erq427. Epub 2011 Jan 31. J Exp Bot. 2011. PMID: 21282331 Review.
-
Nitrate assimilation by bacteria.Adv Microb Physiol. 1998;39:1-30, 379. doi: 10.1016/s0065-2911(08)60014-4. Adv Microb Physiol. 1998. PMID: 9328645 Review.
Cited by
-
Phenotype Assessment and Putative Mechanisms of Ammonium Toxicity to Plants.Int J Mol Sci. 2025 Mar 13;26(6):2606. doi: 10.3390/ijms26062606. Int J Mol Sci. 2025. PMID: 40141246 Free PMC article. Review.
References
-
- Aichi M, Maeda S-I, Ichikawa K, Omata T.. 2004. Nitrite-responsive activation of the nitrate assimilation operon in cyanobacteria plays an essential role in up-regulation of nitrate assimilation activities under nitrate-limited growth conditions. Journal of Bacteriology 186, 3224–3229. - PMC - PubMed
-
- Aichi M, Yoshihara S, Yamashita M, Maeda S, Nagai K, Omata T.. 2006. Characterization of the nitrate-nitrite transporter of the major facilitator superfamily (the nrtP gene product) from the cyanobacterium Nostoc punctiforme strain ATCC 29133. Bioscience, Biotechnology, and Biochemistry 70, 2682–2689. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

