Metabolome dynamics during wheat domestication
- PMID: 35595776
- PMCID: PMC9122938
- DOI: 10.1038/s41598-022-11952-9
Metabolome dynamics during wheat domestication
Abstract
One of the most important crops worldwide is wheat. Wheat domestication took place about 10,000 years ago. Not only that its wild progenitors have been discovered and phenotypically characterized, but their genomes were also sequenced and compared to modern wheat. While comparative genomics is essential to track genes that contribute to improvement in crop yield, comparative analyses of functional biological end-products, such as metabolites, are still lacking. With the advent of rigorous mass-spectrometry technologies, it is now possible to address that problem on a big-data scale. In attempt to reveal classes of metabolites, which are associated with wheat domestication, we analyzed the metabolomes of wheat kernel samples from various wheat lines. These wheat lines represented subspecies of tetraploid wheat along primary and secondary domestications, including wild emmer, domesticated emmer, landraces durum, and modern durum. We detected that the groups of plant metabolites such as plant-defense metabolites, antioxidants and plant hormones underwent significant changes during wheat domestication. Our data suggest that these metabolites may have contributed to the improvement in the agricultural fitness of wheat. Closer evaluation of specific metabolic pathways may result in the future in genetically-engineered high-yield crops.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
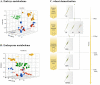
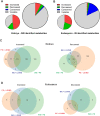
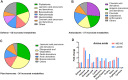
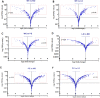
References
MeSH terms
LinkOut - more resources
Full Text Sources

