Assessment of mutations on RBD in the Spike protein of SARS-CoV-2 Alpha, Delta and Omicron variants
- PMID: 35595778
- PMCID: PMC9121086
- DOI: 10.1038/s41598-022-12479-9
Assessment of mutations on RBD in the Spike protein of SARS-CoV-2 Alpha, Delta and Omicron variants
Abstract
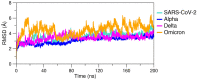
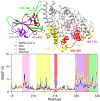
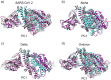
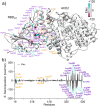
The severe acute respiratory syndrome (SARS) coronavirus 2 (CoV-2) variant Omicron spread more rapid than the other variants of SARS-CoV-2 virus. Mutations on the Spike (S) protein receptor-binding domain (RBD) are critical for the antibody resistance and infectivity of the SARS-CoV-2 variants. In this study, we have used accelerated molecular dynamics (aMD) simulations and free energy calculations to present a systematic analysis of the affinity and conformational dynamics along with the interactions that drive the binding between Spike protein RBD and human angiotensin-converting enzyme 2 (ACE2) receptor. We evaluate the impacts of the key mutation that occur in the RBDs Omicron and other variants in the binding with the human ACE2 receptor. The results show that S protein Omicron has stronger binding to the ACE2 than other variants. The evaluation of the decomposition energy per residue shows the mutations N440K, T478K, Q493R and Q498R observed in Spike protein of SARS-CoV-2 provided a stabilization effect for the interaction between the SARS-CoV-2 RBD and ACE2. Overall, the results demonstrate that faster spreading of SARS-CoV-2 Omicron may be correlated with binding affinity of S protein RBD to ACE2 and mutations of uncharged residues to positively charged residues such as Lys and Arg in key positions in the RBD.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Computational modeling of the effect of five mutations on the structure of the ACE2 receptor and their correlation with infectivity and virulence of some emerged variants of SARS-CoV-2 suggests mechanisms of binding affinity dysregulation.Chem Biol Interact. 2022 Dec 1;368:110244. doi: 10.1016/j.cbi.2022.110244. Epub 2022 Nov 3. Chem Biol Interact. 2022. PMID: 36336003 Free PMC article.
-
V367F Mutation in SARS-CoV-2 Spike RBD Emerging during the Early Transmission Phase Enhances Viral Infectivity through Increased Human ACE2 Receptor Binding Affinity.J Virol. 2021 Jul 26;95(16):e0061721. doi: 10.1128/JVI.00617-21. Epub 2021 Jul 26. J Virol. 2021. PMID: 34105996 Free PMC article.
-
Binding of human ACE2 and RBD of Omicron enhanced by unique interaction patterns among SARS-CoV-2 variants of concern.J Comput Chem. 2023 Feb 5;44(4):594-601. doi: 10.1002/jcc.27025. Epub 2022 Nov 18. J Comput Chem. 2023. PMID: 36398990 Free PMC article.
-
How Do Point Mutations Enhancing the Basic Character of the RBDs of SARS-CoV-2 Variants Affect Their Transmissibility and Infectivity Capacities?Viruses. 2022 Apr 10;14(4):783. doi: 10.3390/v14040783. Viruses. 2022. PMID: 35458513 Free PMC article. Review.
-
Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): a structural perspective.Mol Biol Rep. 2023 Mar;50(3):2713-2721. doi: 10.1007/s11033-022-08193-4. Epub 2022 Dec 23. Mol Biol Rep. 2023. PMID: 36562937 Free PMC article. Review.
Cited by
-
Molecular dynamics simulations highlight the altered binding landscape at the spike-ACE2 interface between the Delta and Omicron variants compared to the SARS-CoV-2 original strain.Comput Biol Med. 2022 Oct;149:106035. doi: 10.1016/j.compbiomed.2022.106035. Epub 2022 Aug 27. Comput Biol Med. 2022. PMID: 36055162 Free PMC article.
-
Aptamer-based diagnosis of various SARS-CoV2 strains isolated from clinical specimens.Heliyon. 2023 Jun;9(6):e16458. doi: 10.1016/j.heliyon.2023.e16458. Epub 2023 May 23. Heliyon. 2023. PMID: 37251485 Free PMC article.
-
Molecular evolution and targeted recombination of SARS-CoV-2 in South Korea.iScience. 2023 Aug 22;26(9):107689. doi: 10.1016/j.isci.2023.107689. eCollection 2023 Sep 15. iScience. 2023. PMID: 37680469 Free PMC article.
-
Research progress of spike protein mutation of SARS-CoV-2 mutant strain and antibody development.Front Immunol. 2024 Nov 18;15:1407149. doi: 10.3389/fimmu.2024.1407149. eCollection 2024. Front Immunol. 2024. PMID: 39624100 Free PMC article. Review.
-
Efficient discovery of frequently co-occurring mutations in a sequence database with matrix factorization.PLoS Comput Biol. 2025 Apr 24;21(4):e1012391. doi: 10.1371/journal.pcbi.1012391. eCollection 2025 Apr. PLoS Comput Biol. 2025. PMID: 40273414 Free PMC article.
References
-
- World Health Organization . Coronavirus Disease (COVID-19) Outbreak. World Health Organization; 2020.
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

