ILDR1 promotes influenza A virus replication through binding to PLSCR1
- PMID: 35595813
- PMCID: PMC9122930
- DOI: 10.1038/s41598-022-12598-3
ILDR1 promotes influenza A virus replication through binding to PLSCR1
Abstract
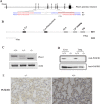
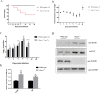
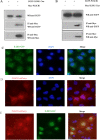
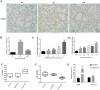
As a natural antiviral regulator, phospholipid scramblase 1 (PLSCR1) has been shown to inhibit influenza virus replication in infected cells through interacting with NP of influenza A virus (IAV). But its antiviral function as well as the underlying regulatory mechanism has not been examined in vivo. In the present work, we show that PLSCR1 expression is decreased in H1N1 SIV-infected mice, and Plscr1-/- mice are more susceptible to H1N1 SIV infection. By performing yeast two-hybrid screening, we identified immunoglobulin-like domain-containing receptor 1 (ILDR1) as a novel PLSCR1-binding partner. ILDR1 is highly expressed in the lungs, and its expression level is increased after virus infection. Interestingly, ILDR1 could not directly interact with virus NP protein, but could combine with PLSCR1 competitively. Our data indicates that there is a previously unidentified PLSCR1-ILDR1-NP regulatory pathway playing a vital role in limiting IAV infection, which provides novel insights into IAV-host interactions.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Phospholipid scramblase 1: a frontline defense against viral infections.Front Cell Infect Microbiol. 2025 Apr 3;15:1573373. doi: 10.3389/fcimb.2025.1573373. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40248364 Free PMC article. Review.
-
Phospholipid Scramblase 1 (PLSCR1) Regulates Interferon-Lambda Receptor 1 (IFN-λR1) and IFN-λ Signaling in Influenza A Virus (IAV) Infection.bioRxiv [Preprint]. 2024 Nov 21:2024.11.20.624469. doi: 10.1101/2024.11.20.624469. bioRxiv. 2024. PMID: 39605457 Free PMC article. Preprint.
-
Phospholipid scramblase 1 interacts with influenza A virus NP, impairing its nuclear import and thereby suppressing virus replication.PLoS Pathog. 2018 Jan 19;14(1):e1006851. doi: 10.1371/journal.ppat.1006851. eCollection 2018 Jan. PLoS Pathog. 2018. PMID: 29352288 Free PMC article.
-
Inhibition effects of novel polyketide compound PPQ-B against influenza A virus replication by interfering with the cellular EGFR pathway.Antiviral Res. 2017 Jul;143:74-84. doi: 10.1016/j.antiviral.2017.04.007. Epub 2017 Apr 13. Antiviral Res. 2017. PMID: 28414053
-
Host Non-Coding RNA Regulates Influenza A Virus Replication.Viruses. 2021 Dec 29;14(1):51. doi: 10.3390/v14010051. Viruses. 2021. PMID: 35062254 Free PMC article. Review.
Cited by
-
Cross one single body 49 tissues single-cell transcriptome reveals detailed macrophage heterogeneity during pig pregnancy.Front Immunol. 2025 Apr 2;16:1574120. doi: 10.3389/fimmu.2025.1574120. eCollection 2025. Front Immunol. 2025. PMID: 40242774 Free PMC article.
-
Phospholipid scramblase 1: a frontline defense against viral infections.Front Cell Infect Microbiol. 2025 Apr 3;15:1573373. doi: 10.3389/fcimb.2025.1573373. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40248364 Free PMC article. Review.
-
Population-enriched innate immune variants may identify candidate gene targets at the intersection of cancer and cardio-metabolic disease.Front Endocrinol (Lausanne). 2024 Mar 21;14:1286979. doi: 10.3389/fendo.2023.1286979. eCollection 2023. Front Endocrinol (Lausanne). 2024. PMID: 38577257 Free PMC article. Review.
-
Phospholipid Scramblase 1 (PLSCR1) Regulates Interferon-Lambda Receptor 1 (IFN-λR1) and IFN-λ Signaling in Influenza A Virus (IAV) Infection.bioRxiv [Preprint]. 2024 Nov 21:2024.11.20.624469. doi: 10.1101/2024.11.20.624469. bioRxiv. 2024. PMID: 39605457 Free PMC article. Preprint.
-
Quantitative proteomics of differentiated primary bronchial epithelial cells from chronic obstructive pulmonary disease and control identifies potential novel host factors post-influenza A virus infection.Front Microbiol. 2023 Jan 11;13:957830. doi: 10.3389/fmicb.2022.957830. eCollection 2022. Front Microbiol. 2023. PMID: 36713229 Free PMC article.
References
-
- Vincent A, Awada L, Brown I, Chen H, Claes F, Dauphin G, Donis R, Culhane M, Hamilton K, Lewis N, Mumford E, Nguyen T, Parchariyanon S, Pasick J, Pavade G, Pereda A, Peiris M, Saito T, Swenson S, Van Reeth K, Webby R, Wong F, Ciacci-Zanella J. Review of influenza A virus in swine worldwide: A call for increased surveillance and research. Zoonoses Public Health. 2014;61(1):4–17. doi: 10.1111/zph.12049. - DOI - PubMed
-
- Sun H, Xiao Y, Liu J, Wang D, Li F, Wang C, Li C, Zhu J, Song J, Sun H, Jiang Z, Liu L, Zhang X, Wei K, Hou D, Pu J, Sun Y, Tong Q, Bi Y, Chang KC, Liu S, Gao GF, Liu J. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc. Natl. Acad. Sci. U S A. 2020;117(29):17204–17210. doi: 10.1073/pnas.1921186117. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

