Ixovex-1, a novel oncolytic E1B-mutated adenovirus
- PMID: 35596069
- PMCID: PMC9663300
- DOI: 10.1038/s41417-022-00480-3
Ixovex-1, a novel oncolytic E1B-mutated adenovirus
Abstract
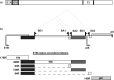
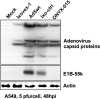
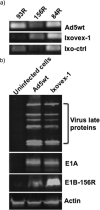
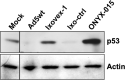
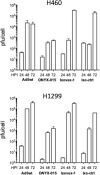
There is a great demand for improved oncolytic viruses that selectively replicate within cancer cells while sparing normal cells. Here, we describe a novel oncolytic adenovirus, Ixovex-1, that obtains a cancer-selective replication phenotype by modulating the level of expression of the different, alternatively spliced E1B mRNA isoforms. Ixovex-1 is a recombinant adenovirus that carries a single point mutation in the E1B-93R 3' splice acceptor site that results in overexpression of the E1B-156R splice isoform. In this paper, we studied the characteristics of this novel oncolytic adenovirus by validating its in vitro behaviour in a panel of normal cells and cancer cells. We additionally studied its anti-tumour efficacy in vivo. Ixovex-1 significantly inhibited tumour growth and prolonged survival of mice in an immune-deficient lung carcinoma tumour implantation model. In complementation experiments, overexpression of E1B-156R was shown to increase the oncolytic index of both Ad5wt and ONYX-015. In contrast to prior viruses of similar type, Ixovex-1 includes a functional E3B region for better in vivo efficacy. Throughout this study, the Ixovex-1 virus has been proven to be superior in competency compared to a virus with multiple deletions.
© 2022. The Author(s).
Conflict of interest statement
AC, GA, M-LA, MR and ME have no conflict of interests. DÖ and MA have both stocks in Ixogen Ltd., UK. GhA is the founder and CEO of Ixogen Ltd. Ixogen holds worldwide patents to Ixovex-1. GhA is also the founder and CEO of PsiVac Ltd., UK.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

