Recombination rates in pigs differ between breeds, sexes and individuals, and are associated with the RNF212, SYCP2, PRDM7, MEI1 and MSH4 loci
- PMID: 35596132
- PMCID: PMC9123673
- DOI: 10.1186/s12711-022-00723-9
Recombination rates in pigs differ between breeds, sexes and individuals, and are associated with the RNF212, SYCP2, PRDM7, MEI1 and MSH4 loci
Abstract
Background: Recombination is a fundamental part of mammalian meiosis that leads to the exchange of large segments of DNA between homologous chromosomes and is therefore an important driver of genetic diversity in populations. In breeding populations, understanding recombination is of particular interest because it can break up unfavourable linkage phases between alleles and produce novel combinations of alleles that could be exploited in selection. In this study, we used dense single nucleotide polymorphism (SNP) genotype data and pedigree information to analyse individual and sex-specific variation and genetic architecture of recombination rates within and between five commercially selected pig breeds.
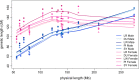
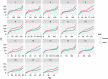
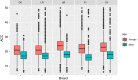
Results: In agreement with previous studies, recombination rates were higher in females than in males for all breeds and for all chromosomes, except 1 and 13, for which male rates were slightly higher. Total recombination rate differed between breeds but the pattern of recombination along the chromosomes was well conserved across breeds for the same sex. The autosomal linkage maps spanned a total length of 1731 to 1887 cM for males and of 2231 to 2515 cM for females. Estimates of heritability for individual autosomal crossover count ranged from 0.04 to 0.07 for males and from 0.08 to 0.11 for females. Fourteen genomic regions were found to be associated with individual autosomal crossover count. Of these, four were close to or within candidate genes that have previously been associated with individual recombination rates in pigs and other mammals, namely RNF212, SYCP2 and MSH4. Two of the identified regions included the PRDM7 and MEI1 genes, which are known to be involved in meiosis but have not been previously associated with variation in individual recombination rates.
Conclusions: This study shows that genetic variation in autosomal recombination rate persists in domesticated species under strong selection, with differences between closely-related breeds and marked differences between the sexes. Our findings support results from other studies, i.e., that individual crossover counts are associated with the RNF212, SYCP2 and MSH4 genes in pig. In addition, we have found two novel candidate genes associated with the trait, namely PRDM7 and MEI1.
© 2022. The Author(s).
Conflict of interest statement
AG is employed in Topigs Norsvin. All authors declare that the results are presented in full and as such present no conflict of interest. The other authors declare that they have no competing interests for this study.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

