Machine learning and comparative genomics approaches for the discovery of xylose transporters in yeast
- PMID: 35596177
- PMCID: PMC9123741
- DOI: 10.1186/s13068-022-02153-7
Machine learning and comparative genomics approaches for the discovery of xylose transporters in yeast
Abstract
Background: The need to mitigate and substitute the use of fossil fuels as the main energy matrix has led to the study and development of biofuels as an alternative. Second-generation (2G) ethanol arises as one biofuel with great potential, due to not only maintaining food security, but also as a product from economically interesting crops such as energy-cane. One of the main challenges of 2G ethanol is the inefficient uptake of pentose sugars by industrial yeast Saccharomyces cerevisiae, the main organism used for ethanol production. Understanding the main drivers for xylose assimilation and identify novel and efficient transporters is a key step to make the 2G process economically viable.
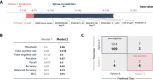
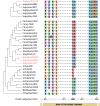
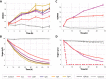
Results: By implementing a strategy of searching for present motifs that may be responsible for xylose transport and past adaptations of sugar transporters in xylose fermenting species, we obtained a classifying model which was successfully used to select four different candidate transporters for evaluation in the S. cerevisiae hxt-null strain, EBY.VW4000, harbouring the xylose consumption pathway. Yeast cells expressing the transporters SpX, SpH and SpG showed a superior uptake performance in xylose compared to traditional literature control Gxf1.
Conclusions: Modelling xylose transport with the small data available for yeast and bacteria proved a challenge that was overcome through different statistical strategies. Through this strategy, we present four novel xylose transporters which expands the repertoire of candidates targeting yeast genetic engineering for industrial fermentation. The repeated use of the model for characterizing new transporters will be useful both into finding the best candidates for industrial utilization and to increase the model's predictive capabilities.
Keywords: Feature selection; Industrial biotechnology; Machine learning; Pentose metabolism; Xylose; Xylose transporter.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Dias MOS, Junqueira TL, Cavalett O, Pavanello LG, Cunha MP, Jesus CDF, et al. Biorefineries for the production of first and second generation ethanol and electricity from sugarcane. App Energy. 2013;109:72–78. doi: 10.1016/j.apenergy.2013.03.081. - DOI
-
- Balat M. Production of bioethanol from lignocellulosic materials via the biochemical pathway : a review. Energy Convers Manag. 2011;52:858–875. doi: 10.1016/j.enconman.2010.08.013. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
