Physiological and comparative transcriptome analyses reveal the mechanisms underlying waterlogging tolerance in a rapeseed anthocyanin-more mutant
- PMID: 35596185
- PMCID: PMC9123723
- DOI: 10.1186/s13068-022-02155-5
Physiological and comparative transcriptome analyses reveal the mechanisms underlying waterlogging tolerance in a rapeseed anthocyanin-more mutant
Abstract
Background: Rapeseed (Brassica napus) is the second largest oil crop worldwide. It is widely used in food, energy production and the chemical industry, as well as being an ornamental. Consequently, it has a large economic value and developmental potential. Waterlogging is an important abiotic stress that restricts plant growth and development. However, little is known about the molecular mechanisms underlying waterlogging tolerance in B. napus.
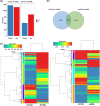
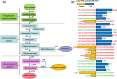
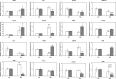
Results: In the present study, the physiological changes and transcriptomes of germination-stage rapeseed in response to waterlogging stress were investigated in the B. napus cultivar 'Zhongshuang 11' (ZS11) and its anthocyanin-more (am) mutant, which was identified in our previous study. The mutant showed stronger waterlogging tolerance compared with ZS11, and waterlogging stress significantly increased anthocyanin, soluble sugar and malondialdehyde contents and decreased chlorophyll contents in the mutant after 12 days of waterlogging. An RNA-seq analysis identified 1370 and 2336 differently expressed genes (DEGs) responding to waterlogging stress in ZS11 and am, respectively. An enrichment analysis revealed that the DEGs in ZS11 were predominately involved in carbohydrate metabolism, whereas those in the am mutant were particularly enriched in plant hormone signal transduction and response to endogenous stimulation. In total, 299 DEGs were identified as anthocyanin biosynthesis-related structural genes (24) and regulatory genes encoding transcription factors (275), which may explain the increased anthocyanin content in the am mutant. A total of 110 genes clustered in the plant hormone signal transduction pathway were also identified as DEGs, including 70 involved in auxin and ethylene signal transduction that were significantly changed in the mutant. Furthermore, the expression levels of 16 DEGs with putative roles in anthocyanin accumulation and biotic/abiotic stress responses were validated by quantitative real-time PCR as being consistent with the transcriptome profiles.
Conclusion: This study provides new insights into the molecular mechanisms of increased anthocyanin contents in rapeseed in response to waterlogging stress, which should be useful for reducing the damage caused by waterlogging stress and for further breeding new rapeseed varieties with high waterlogging tolerance.
Keywords: Anthocyanin; Brassica napus; Candidate gene; Physiological; Transcriptome; Waterlogging stress.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Pullen J, Saeed K. Investigation of the factors affecting the progress of base-catalyzed transesterification of rapeseed oil to biodiesel FAME. Fuel Process Technol. 2015;130:127–135.
-
- Ding LN, Li T, Guo XJ, Li M, Liu XY, Cao J, et al. Sclerotinia stem rot resistance in rapeseed: recent progress and future prospects. J Agric Food Chem. 2021;69:2965–2978. - PubMed
Grants and funding
- 32172060 and 31471527/National Natural Science Foundation of China
- 32172060 and 31471527/National Natural Science Foundation of China
- BK20201416)/Natural Science Foundation of Jiangsu Province
- CX(21)3112/Jiangsu Agriculture science and technology innovation fund
- 2016YFD0101904 and 2016YFD0100305/National Key R & D Program of China
LinkOut - more resources
Full Text Sources
