Polyploid giant cancer cells, EZH2 and Myc upregulation in mammary epithelial cells infected with high-risk human cytomegalovirus
- PMID: 35596973
- PMCID: PMC9121245
- DOI: 10.1016/j.ebiom.2022.104056
Polyploid giant cancer cells, EZH2 and Myc upregulation in mammary epithelial cells infected with high-risk human cytomegalovirus
Abstract
Background: Human cytomegalovirus (HCMV) infection has been actively implicated in complex neoplastic processes. Beyond oncomodulation, the molecular mechanisms that might underlie HCMV-induced oncogenesis are being extensively studied. Polycomb repressive complex 2 (PRC2) proteins, in particular enhancer of zeste homolog 2 (EZH2) are associated with cancer progression. Nevertheless, little is known about EZH2 activation in the context of HCMV infection and breast oncogenesis.
Methods: Herein, we identified EZH2 as a downstream target for HCMV-induced Myc upregulation upon acute and chronic infection with high-risk strains using a human mammary epithelial model.
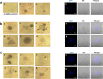
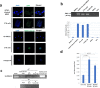
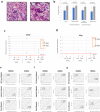
Findings: We detected polyploidy and CMV-transformed HMECs (CTH) cells harboring HCMV and dynamically undergoing the giant cells cycle. Acquisition of embryonic stemness markers positively correlated with EZH2 and Myc expression. EZH2 inhibitors curtail sustained CTH cells' malignant phenotype. Besides harboring polyploid giant cancer cells (PGCCs), tumorigenic breast biopsies were characterized by an enhanced EZH2 and Myc expression, with a strong positive correlation between EZH2 and Myc expression, and between PGCC count and EZH2/Myc expression in the presence of HCMV. Further, we isolated two HCMV strains from EZH2HighMycHigh basal-like tumors which replicate in MRC5 cells and transform HMECs toward CTH cells after acute infection.
Interpretation: Our data establish a potential link between HCMV-induced Myc activation, the subsequent EZH2 upregulation, and polyploidy induction. These data support the proposed tumorigenesis properties of EZH2/Myc, and allow the isolation of two oncogenic HCMV strains from EZH2HighMycHigh basal breast tumors while identifying EZH2 as a potential therapeutic target in the management of breast cancer, particularly upon HCMV infection.
Funding: This work was supported by grants from the University of Franche-Comté (UFC) (CR3300), the Région Franche-Comté (2021-Y-08292 and 2021-Y-08290) and the Ligue contre le Cancer (CR3304) to Georges Herbein. Zeina Nehme is a recipient of a doctoral scholarship from the municipality of Habbouch. Sandy Haidar Ahmad is recipient of a doctoral scholarship from Lebanese municipality. Ranim El Baba is a recipient of a doctoral scholarship from Hariri foundation for sustainable human development.
Keywords: Breast cancer; CTH cells; Cytomegalovirus; EZH2; Myc; Polyploid giant cancer cells.
Copyright © 2022 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures