Complex and unexpected outcomes of antibiotic therapy against a polymicrobial infection
- PMID: 35597889
- PMCID: PMC9381758
- DOI: 10.1038/s41396-022-01252-5
Complex and unexpected outcomes of antibiotic therapy against a polymicrobial infection
Abstract
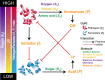
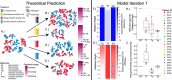
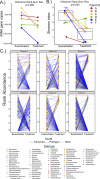
Antibiotics are our primary approach to treating complex infections, yet we have a poor understanding of how these drugs affect microbial communities. To better understand antimicrobial effects on host-associated microbial communities we treated cultured sputum microbiomes from people with cystic fibrosis (pwCF, n = 24) with 11 different antibiotics, supported by theoretical and mathematical modeling-based predictions in a mucus-plugged bronchiole microcosm. Treatment outcomes we identified in vitro that were predicted in silico were: 1) community death, 2) community resistance, 3) pathogen killing, and 4) fermenter killing. However, two outcomes that were not predicted when antibiotics were applied were 5) community profile shifts with little change in total bacterial load (TBL), and 6) increases in TBL. The latter outcome was observed in 17.8% of samples with a TBL increase of greater than 20% and 6.8% of samples with an increase greater than 40%, demonstrating significant increases in community carrying capacity in the presence of an antibiotic. An iteration of the mathematical model showed that TBL increase was due to antibiotic-mediated release of pH-dependent inhibition of pathogens by anaerobe fermentation. These dynamics were verified in vitro when killing of fermenters resulted in a higher community carrying capacity compared to a no antibiotic control. Metagenomic sequencing of sputum samples during antibiotic therapy revealed similar dynamics in clinical samples. This study shows that the complex microbial ecology dictates the outcomes of antibiotic therapy against a polymicrobial infection.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

