Exome-wide analysis of copy number variation shows association of the human leukocyte antigen region with asthma in UK Biobank
- PMID: 35597955
- PMCID: PMC9124406
- DOI: 10.1186/s12920-022-01268-y
Exome-wide analysis of copy number variation shows association of the human leukocyte antigen region with asthma in UK Biobank
Abstract
Background: The role of copy number variants (CNVs) in susceptibility to asthma is not well understood. This is, in part, due to the difficulty of accurately measuring CNVs in large enough sample sizes to detect associations. The recent availability of whole-exome sequencing (WES) in large biobank studies provides an unprecedented opportunity to study the role of CNVs in asthma.
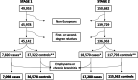
Methods: We called common CNVs in 49,953 individuals in the first release of UK Biobank WES using ClinCNV software. CNVs were tested for association with asthma in a stage 1 analysis comprising 7098 asthma cases and 36,578 controls from the first release of sequencing data. Nominally-associated CNVs were then meta-analysed in stage 2 with an additional 17,280 asthma cases and 115,562 controls from the second release of UK Biobank exome sequencing, followed by validation and fine-mapping.
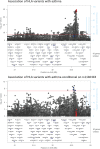
Results: Five of 189 CNVs were associated with asthma in stage 2, including a deletion overlapping the HLA-DQA1 and HLA-DQB1 genes, a duplication of CHROMR/PRKRA, deletions within MUC22 and TAP2, and a duplication in FBRSL1. The HLA-DQA1, HLA-DQB1, MUC22 and TAP2 genes all reside within the human leukocyte antigen (HLA) region on chromosome 6. In silico analyses demonstrated that the deletion overlapping HLA-DQA1 and HLA-DQB1 is likely to be an artefact arising from under-mapping of reads from non-reference HLA haplotypes, and that the CHROMR/PRKRA and FBRSL1 duplications represent presence/absence of pseudogenes within the HLA region. Bayesian fine-mapping of the HLA region suggested that there are two independent asthma association signals. The variants with the largest posterior inclusion probability in the two credible sets were an amino acid change in HLA-DQB1 (glutamine to histidine at residue 253) and a multi-allelic amino acid change in HLA-DRB1 (presence/absence of serine, glycine or leucine at residue 11).
Conclusions: At least two independent loci characterised by amino acid changes in the HLA-DQA1, HLA-DQB1 and HLA-DRB1 genes are likely to account for association of SNPs and CNVs in this region with asthma. The high divergence of haplotypes in the HLA can give rise to spurious CNVs, providing an important, cautionary tale for future large-scale analyses of sequencing data.
Keywords: Asthma; Copy number variants; Exome sequencing; Fine-mapping; Genetic association; Human leukocyte antigen; UK Biobank.
© 2022. The Author(s).
Conflict of interest statement
LVW has research funding (outside of submitted work) from GSK and Orion Pharma and consultancy for Galapagos. All other authors declare that they have no competing interests.
Figures
Similar articles
-
HLA class I (A, B, C) and class II (DRB1, DQA1, DQB1, DPB1) alleles and haplotypes in the Han from southern China.Tissue Antigens. 2007 Dec;70(6):455-63. doi: 10.1111/j.1399-0039.2007.00932.x. Epub 2007 Sep 27. Tissue Antigens. 2007. PMID: 17900288
-
Association of HLA-DRB1 and -DQ Alleles and Haplotypes with Type 1 Diabetes in Jordanians.Endocr Metab Immune Disord Drug Targets. 2020;20(6):895-902. doi: 10.2174/1871530319666191119114031. Endocr Metab Immune Disord Drug Targets. 2020. PMID: 31742498
-
Association of insulin-dependent diabetes mellitus in Taiwan with HLA class II DQB1 and DRB1 alleles.Hum Immunol. 1993 Oct;38(2):105-14. doi: 10.1016/0198-8859(93)90526-7. Hum Immunol. 1993. PMID: 8106265
-
DRB1-DQA1-DQB1 loci and multiple sclerosis predisposition in the Sardinian population.Hum Mol Genet. 1998 Aug;7(8):1235-7. doi: 10.1093/hmg/7.8.1235. Hum Mol Genet. 1998. PMID: 9668164
-
Genetic Susceptibility to Chronic Kidney Disease - Some More Pieces for the Heritability Puzzle.Front Genet. 2019 May 31;10:453. doi: 10.3389/fgene.2019.00453. eCollection 2019. Front Genet. 2019. PMID: 31214239 Free PMC article. Review.
Cited by
-
Titin copy number variations associated with dominant inherited phenotypes.J Med Genet. 2024 Mar 21;61(4):369-377. doi: 10.1136/jmg-2023-109473. J Med Genet. 2024. PMID: 37935568 Free PMC article.
-
CNest: A novel copy number association discovery method uncovers 862 new associations from 200,629 whole-exome sequence datasets in the UK Biobank.Cell Genom. 2022 Aug 10;2(8):100167. doi: 10.1016/j.xgen.2022.100167. eCollection 2022 Aug 10. Cell Genom. 2022. PMID: 36779085 Free PMC article.
-
The Multifaceted Roles of CHROMR in Innate Immunity, Cancer, and Cholesterol Homeostasis.Noncoding RNA. 2025 Jun 10;11(3):44. doi: 10.3390/ncrna11030044. Noncoding RNA. 2025. PMID: 40559622 Free PMC article. Review.
-
Asthma-Genomic Advances Toward Risk Prediction.Clin Chest Med. 2024 Sep;45(3):599-610. doi: 10.1016/j.ccm.2024.03.002. Epub 2024 Apr 21. Clin Chest Med. 2024. PMID: 39069324 Review.
-
Copy Number Variation in Asthma: An Integrative Review.Clin Rev Allergy Immunol. 2025 Jan 4;68(1):4. doi: 10.1007/s12016-024-09015-0. Clin Rev Allergy Immunol. 2025. PMID: 39755867 Review.
References
-
- Oliveira P, Costa GNO, Damasceno AKA, Hartwig FP, Barbosa GCG, Figueiredo CA, et al. Genome-wide burden and association analyses implicate copy number variations in asthma risk among children and young adults from Latin America. Sci Rep. 2018;8(1):14475. doi: 10.1038/s41598-018-32837-w. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

