Whole-genome sequencing uncovers the structural and transcriptomic landscape of hexaploid wheat/Ambylopyrum muticum introgression lines
- PMID: 35598169
- PMCID: PMC9946142
- DOI: 10.1111/pbi.13859
Whole-genome sequencing uncovers the structural and transcriptomic landscape of hexaploid wheat/Ambylopyrum muticum introgression lines
Abstract
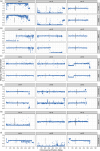
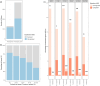
Wheat is a globally vital crop, but its limited genetic variation creates a challenge for breeders aiming to maintain or accelerate agricultural improvements over time. Introducing novel genes and alleles from wheat's wild relatives into the wheat breeding pool via introgression lines is an important component of overcoming this low variation but is constrained by poor genomic resolution and limited understanding of the genomic impact of introgression breeding programmes. By sequencing 17 hexaploid wheat/Ambylopyrum muticum introgression lines and the parent lines, we have precisely pinpointed the borders of introgressed segments, most of which occur within genes. We report a genome assembly and annotation of Am. muticum that has facilitated the identification of Am. muticum resistance genes commonly introgressed in lines resistant to stripe rust. Our analysis has identified an abundance of structural disruption and homoeologous pairing across the introgression lines, likely caused by the suppressed Ph1 locus. mRNAseq analysis of six of these introgression lines revealed that novel introgressed genes are rarely expressed and those that directly replace a wheat orthologue have a tendency towards downregulation, with no discernible compensation in the expression of homoeologous copies. This study explores the genomic impact of introgression breeding and provides a schematic that can be followed to characterize introgression lines and identify segments and candidate genes underlying the phenotype. This will facilitate more effective utilization of introgression pre-breeding material in wheat breeding programmes.
Keywords: Wheat; breeding; genomics; introgression; resistance; wild relative.
© 2022 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Alexa, A. , Rahnenfuhrer, J. , (2021) topGO: enrichment analysis for gene ontology. R package version 2.38.1.
-
- Charmet, G. (2011) Wheat domestication: lessons for the future. C. R. Biol. 334, 212–220. - PubMed
-
- Chen, S. , Rouse, M.N. , Zhang, W. , Zhang, X. , Guo, Y. , Briggs, J. and Dubcovsky, J. (2020) Wheat gene Sr60 encodes a protein with two putative kinase domains that confers resistance to stem rust. New Phytol. 225, 948–959. - PubMed
Publication types
MeSH terms
Grants and funding
- BB/CCG1720/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M011216/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P016855/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J004596/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

