Overexpression of a Zea mays Brassinosteroid-Signaling Kinase Gene ZmBSK1 Confers Salt Stress Tolerance in Maize
- PMID: 35599886
- PMCID: PMC9121125
- DOI: 10.3389/fpls.2022.894710
Overexpression of a Zea mays Brassinosteroid-Signaling Kinase Gene ZmBSK1 Confers Salt Stress Tolerance in Maize
Abstract
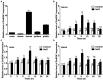
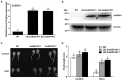
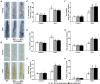
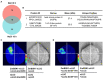
Salinity has become a crucial environmental factor seriously restricting maize (Zea mays L.) growth, development and productivity. However, how plants respond to salt stress is still poorly understood. In this study, we report that a maize brassinosteroid-signaling kinase gene ZmBSK1 plays a significant role in salt stress response. Expression pattern analysis revealed that the transcript level of ZmBSK1 was upregulated by NaCl treatment both in maize leaves, roots, and stems. Phenotypic and physiological analysis showed that overexpression of ZmBSK1 in maize improved salt tolerance by reducing the malondialdehyde (MDA) content, the percentage of electrolyte leakage, O2 - and H2O2 accumulation under salt stress, relying on the increases of antioxidant defense enzyme activities and proline content. qRT-PCR analysis showed that overexpression of ZmBSK1 also positively modulated the expression levels of reactive oxygen species (ROS)-scavenging and proline biosynthesis-related genes under salt stress. Moreover, immunoprecipitation-mass spectrometry (IP-MS) assay and firefly luciferase complementation imaging (LCI) assay showed that ZmBSK1 could associate with heat shock protein ZmHSP8 and 14-3-3-like protein ZmGF14-6, and their gene expression levels could be significantly induced by NaCl treatment in different maize tissues. Our findings unravel the new function of ZmBSK1 in salt stress response, which provides the theoretical bases for the improvement of maize salt resistance.
Keywords: ZmBSK1; antioxidant defense enzyme; maize; protein interaction; reactive oxygen species; salt tolerance.
Copyright © 2022 Liu, Sun, Di, Cui, Meng, Wu, Chen and Yuan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
ZmBSK1 positively regulates BR-induced H2O2 production via NADPH oxidase and functions in oxidative stress tolerance in maize.Plant Physiol Biochem. 2022 Aug 15;185:325-335. doi: 10.1016/j.plaphy.2022.06.011. Epub 2022 Jun 14. Plant Physiol Biochem. 2022. PMID: 35738188
-
BRASSINOSTEROID-SIGNALING KINASE 1 phosphorylating CALCIUM/CALMODULIN-DEPENDENT PROTEIN KINASE functions in drought tolerance in maize.New Phytol. 2021 Jul;231(2):695-712. doi: 10.1111/nph.17403. Epub 2021 May 17. New Phytol. 2021. PMID: 33864702
-
Brassinosteroid-signaling kinase ZmBSK7 enhances salt stress tolerance in maize.Biochem Biophys Res Commun. 2024 Sep 3;723:150222. doi: 10.1016/j.bbrc.2024.150222. Epub 2024 Jun 3. Biochem Biophys Res Commun. 2024. PMID: 38850813
-
Interruption of Jasmonic Acid Biosynthesis Causes Differential Responses in the Roots and Shoots of Maize Seedlings against Salt Stress.Int J Mol Sci. 2019 Dec 9;20(24):6202. doi: 10.3390/ijms20246202. Int J Mol Sci. 2019. PMID: 31835299 Free PMC article.
-
Research Progress on the Mechanism of Salt Tolerance in Maize: A Classic Field That Needs New Efforts.Plants (Basel). 2023 Jun 18;12(12):2356. doi: 10.3390/plants12122356. Plants (Basel). 2023. PMID: 37375981 Free PMC article. Review.
Cited by
-
GmBSK1-GmGSK1-GmBES1.5 regulatory module controls heat tolerance in soybean.J Adv Res. 2025 Jul;73:187-198. doi: 10.1016/j.jare.2024.09.004. Epub 2024 Sep 3. J Adv Res. 2025. PMID: 39236976 Free PMC article.
-
Cloning and expression study of a high-affinity nitrate transporter gene from Zea mays L.Plant Signal Behav. 2023 Dec 31;18(1):2163342. doi: 10.1080/15592324.2022.2163342. Plant Signal Behav. 2023. PMID: 36645908 Free PMC article.
-
Drought-tolerant fungal microbes, Aspergillus oryzae and Aspergillus fumigatus, elevate physiohormonal and antioxidant responses of maize under drought stress.Front Microbiol. 2024 Nov 28;15:1488639. doi: 10.3389/fmicb.2024.1488639. eCollection 2024. Front Microbiol. 2024. PMID: 39669778 Free PMC article.
-
Interactions between Brassinosteroids and Strigolactones in Alleviating Salt Stress in Maize.Int J Mol Sci. 2024 Sep 29;25(19):10505. doi: 10.3390/ijms251910505. Int J Mol Sci. 2024. PMID: 39408841 Free PMC article.
-
Identification, evolution, and expression analysis of OsBSK gene family in Oryza sativa Japonica.BMC Plant Biol. 2022 Dec 5;22(1):565. doi: 10.1186/s12870-022-03905-1. BMC Plant Biol. 2022. PMID: 36464674 Free PMC article.
References
-
- Blein-Nicolas M., Negro S. S., Balliau T., Welcker C., Cabrera-Bosquet L., Nicolas S. D., et al. . (2020). A systems genetics approach reveals environment-dependent associations between SNPs, protein coexpression, and drought-related traits in maize. Genome Res. 30, 1593–1604. doi: 10.1101/gr.255224.119, PMID: - DOI - PMC - PubMed
-
- Campo S., Peris-Peris C., Montesinos L., Peñas G., Messeguer J., San Segundo B. (2012). Expression of the maize ZmGF14-6 gene in rice confers tolerance to drought stress while enhancing susceptibility to pathogen infection. J. Exp. Bot. 63, 983–999. doi: 10.1093/jxb/err328, PMID: - DOI - PMC - PubMed
-
- Chen G., Wang Y. Y., Wang X. L., Yang Q., Quan X. Y., Zeng J. B., et al. . (2019a). Leaf epidermis transcriptome reveals drought-induced hormonal signaling for stomatal regulation in wild barley. Plant Growth Regul. 87, 39–54. doi: 10.1007/s10725-018-0450-0 - DOI
LinkOut - more resources
Full Text Sources

