Comparative Transcriptomics Analysis of the Symbiotic Germination of D. officinale (Orchidaceae) With Emphasis on Plant Cell Wall Modification and Cell Wall-Degrading Enzymes
- PMID: 35599894
- PMCID: PMC9120867
- DOI: 10.3389/fpls.2022.880600
Comparative Transcriptomics Analysis of the Symbiotic Germination of D. officinale (Orchidaceae) With Emphasis on Plant Cell Wall Modification and Cell Wall-Degrading Enzymes
Abstract
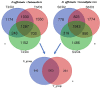
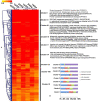
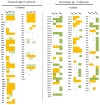
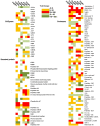
Orchid seed germination in nature is an extremely complex physiological and ecological process involving seed development and mutualistic interactions with a restricted range of compatible mycorrhizal fungi. The impact of the fungal species' partner on the orchids' transcriptomic and metabolic response is still unknown. In this study, we performed a comparative transcriptomic analysis between symbiotic and asymbiotic germination at three developmental stages based on two distinct fungi (Tulasnella sp. and Serendipita sp.) inoculated to the same host plant, Dendrobium officinale. Differentially expressed genes (DEGs) encoding important structural proteins of the host plant cell wall were identified, such as epidermis-specific secreted glycoprotein, proline-rich receptor-like protein, and leucine-rich repeat (LRR) extensin-like protein. These DEGs were significantly upregulated in the symbiotic germination stages and especially in the protocorm stage (stage 3) and seedling stage (stage 4). Differentially expressed carbohydrate-active enzymes (CAZymes) in symbiotic fungal mycelium were observed, they represented 66 out of the 266 and 99 out of the 270 CAZymes annotated in Tulasnella sp. and Serendipita sp., respectively. These genes were speculated to be involved in the reduction of plant immune response, successful colonization by fungi, or recognition of mycorrhizal fungi during symbiotic germination of orchid seed. Our study provides important data to further explore the molecular mechanism of symbiotic germination and orchid mycorrhiza and contribute to a better understanding of orchid seed biology.
Keywords: CAZymes; Serendipita sp.; Tulasnella sp.; comparative transcriptome; symbiotic germination.
Copyright © 2022 Chen, Tang, Kohler, Lebreton, Xing, Zhou, Li, Martin and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

