Evolution of the SARS-CoV-2 pandemic in India
- PMID: 35599988
- PMCID: PMC9108071
- DOI: 10.1016/j.mjafi.2022.05.006
Evolution of the SARS-CoV-2 pandemic in India
Abstract
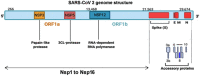
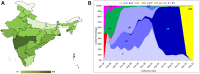
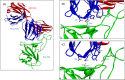
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a bat-derived betacoronavirus, that emerged around December 2019. In spite of the lesser genomic diversity of CoVs in general, a steady accumulation of mutations spread over its genome have been noted, resulting in the emergence of several clades and lineages. Majority of these mutations are random and non-functional changes; however a few variants of concern (VOC) and variants of interest (VOI) designated by the WHO since late 2020 have implications to diagnostics, pathogenicity and immune escape. This review discusses the various nomenclatures depicting the SARS-CoV-2 evolution, the designated VOCs and VOIs and the mutations characterizing these variants. The evolution of SARS-CoV-2 in India and the implications to vaccine efficacy and breakthrough infections is also addressed.
Keywords: Evolution; Genome; Mutations; SARS-CoV-2; Variants of concern.
© 2022 Director General, Armed Forces Medical Services. Published by Elsevier, a division of RELX India Pvt. Ltd.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

