Characterization of sulfated polysaccharide activity against virulent Plasmodium falciparum PHISTb/RLP1 protein
- PMID: 35600144
- PMCID: PMC9096147
- DOI: 10.12688/f1000research.26756.2
Characterization of sulfated polysaccharide activity against virulent Plasmodium falciparum PHISTb/RLP1 protein
Abstract
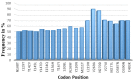
Background: The emergence of artemisinin resistance in South East Asia calls for urgent discovery of new drug compounds that have antiplasmodial activity. Unlike the classical compound screening drug discovery methods, the rational approach involving targeted drug discovery is less cumbersome and therefore key for innovation of new antiplasmodial compounds. Plasmodium falciparum (Pf) utilizes the process of host erythrocyte remodeling using Plasmodium-helical interspersed sub-telomeric domain (PHIST) containing proteins, which are amenable drug targets. The aim of this study is to identify inhibitors of PHIST from sulfated polysaccharides as new antimalarials. Methods: 251 samples from an ongoing study of epidemiology of malaria and drug resistance sensitivity patterns in Kenya were sequenced for PHISTb/RLP1 gene using Sanger sequencing. The sequenced reads were mapped to the reference Pf3D7 protein sequence of PHISTb/RLP1 using CLC Main Workbench. Homology modeling of both reference and mutant protein structures was achieved using the LOMETs tool. The models were refined using ModRefiner for energy minimization. Ramachandran plot was generated by ProCheck to assess the conformation of amino acids in the protein model. Protein binding sites predictions were assessed using FT SITE software. We searched for prospective antimalarials from PubChem. Docking experiments were achieved using AutoDock Vina and analysis results visualized in PyMOL. Results: Sanger sequencing generated 86 complete sequences. Upon mapping of the sequences to the reference, 12 non-synonymous single nucleotide polymorphisms were considered for mutant protein structure analysis. Eleven drug compounds with antiplasmodial activity were identified. Both modeled PHISTb/RLP1 reference and mutant structures had a Ramachandran score of >90% of the amino acids in the favored region. Ten of the drug compounds interacted with amino acid residues in PHISTb and RESA domains, showing potential activity against these proteins. Conclusion: This research identifies inhibitors of exported proteins that can be used in in vitro tests against the Plasmodium parasite.
Keywords: Anti-malarials; Exported proteins; Interactions; P. falciparum; PHISTb/RLP1; Sulfated polysaccharides.
Copyright: © 2022 Mutisya JM et al.
Conflict of interest statement
No competing interests were disclosed.
Figures




References
-
- Alzohairy A: (3) Building a Multiple Sequence Alignment (8) Jalview Basics in More Details.2014. 10.1007/s00436-005-1426-3 - DOI
-
- AutoDock Vina. . Reference Source
Grants and funding
LinkOut - more resources
Full Text Sources

