Highly multiplexed immune repertoire sequencing links multiple lymphocyte classes with severity of response to COVID-19
- PMID: 35600330
- PMCID: PMC9106482
- DOI: 10.1016/j.eclinm.2022.101438
Highly multiplexed immune repertoire sequencing links multiple lymphocyte classes with severity of response to COVID-19
Abstract
Background: Disease progression of subjects with coronavirus disease 2019 (COVID-19) varies dramatically. Understanding the various types of immune response to SARS-CoV-2 is critical for better clinical management of coronavirus outbreaks and to potentially improve future therapies. Disease dynamics can be characterized by deciphering the adaptive immune response.
Methods: In this cross-sectional study we analyzed 117 peripheral blood immune repertoires from healthy controls and subjects with mild to severe COVID-19 disease to elucidate the interplay between B and T cells. We used an immune repertoire Primer Extension Target Enrichment method (immunoPETE) to sequence simultaneously human leukocyte antigen (HLA) restricted T cell receptor beta chain (TRB) and unrestricted T cell receptor delta chain (TRD) and immunoglobulin heavy chain (IgH) immune receptor repertoires. The distribution was analyzed of TRB, TRD and IgH clones between healthy and COVID-19 infected subjects. Using McFadden's Adjusted R2 variables were examined for a predictive model. The aim of this study is to analyze the influence of the adaptive immune repertoire on the severity of the disease (value on the World Health Organization Clinical Progression Scale) in COVID-19.
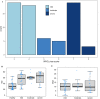
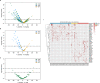
Findings: Combining clinical metadata with clonotypes of three immune receptor heavy chains (TRB, TRD, and IgH), we found significant associations between COVID-19 disease severity groups and immune receptor sequences of B and T cell compartments. Logistic regression showed an increase in shared IgH clonal types and decrease of TRD in subjects with severe COVID-19. The probability of finding shared clones of TRD clonal types was highest in healthy subjects (controls). Some specific TRB clones seems to be present in severe COVID-19 (Figure S7b). The most informative models (McFadden´s Adjusted R2=0.141) linked disease severity with immune repertoire measures across all three cell types, as well as receptor-specific cell counts, highlighting the importance of multiple lymphocyte classes in disease progression.
Interpretation: Adaptive immune receptor peripheral blood repertoire measures are associated with COVID-19 disease severity.
Funding: The study was funded with grants from the Berlin Institute of Health (BIH).
Keywords: COVID-19; Clinical course; Immune receptor; Immune repertoires.
© 2022 The Authors.
Conflict of interest statement
Telman Dilduz, Dannenbaum Richard, Anja Blüher, Florian Rubelt, Gracie Du Zhipei, Luong Khai, Asgharin Hosseinali, Lin Hai and Berka Jan are employees of Roche Diagnostics and Dannenbaum Richard, Rubelt Forian, Lin Hai, Luong Khai, Berka Jan receive salary, stock and options as part of their employment compensation.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

