FAS2FURIOUS: Moderate-Throughput Secreted Expression of Difficult Recombinant Proteins in Drosophila S2 Cells
- PMID: 35600892
- PMCID: PMC9117644
- DOI: 10.3389/fbioe.2022.871933
FAS2FURIOUS: Moderate-Throughput Secreted Expression of Difficult Recombinant Proteins in Drosophila S2 Cells
Abstract
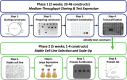
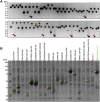
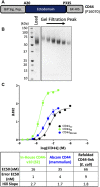
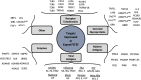
Recombinant protein expression in eukaryotic insect cells is a powerful approach for producing challenging targets. However, due to incompatibility with standard baculoviral platforms and existing low-throughput methodology, the use of the Drosophila melanogaster "S2" cell line lags behind more common insect cell lines such as Sf9 or High-Five™. Due to the advantages of S2 cells, particularly for secreted and secretable proteins, the lack of a simple and parallelizable S2-based platform represents a bottleneck, particularly for biochemical and biophysical laboratories. Therefore, we developed FAS2FURIOUS, a simple and rapid S2 expression pipeline built upon an existing low-throughput commercial platform. FAS2FURIOUS is comparable in effort to simple E. coli systems and allows users to clone and test up to 46 constructs in just 2 weeks. Given the ability of S2 cells to express challenging targets, including receptor ectodomains, secreted glycoproteins, and viral antigens, FAS2FURIOUS represents an attractive orthogonal approach for protein expression in eukaryotic cells.
Keywords: S2 cells; glycoproteins; insect cells; recombinant proteins; secreted proteins; structural biology.
Copyright © 2022 Coker, Katis, Fairhead, Schwenzer, Clemmensen, Frandsen, de Jongh, Gileadi, Burgess-Brown, Marsden, Midwood and Yue.
Conflict of interest statement
SC, BF, and WdJ were employed by ExpreS2ion Biotechnologies, which markets the ExpreS2ion system that was used as a starting point to create FAS2FURIOUS. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Adriaan de Jongh W., Salgueiro S., Dyring C. (2013). The Use ofDrosophilaS2 Cells in R&D and Bioprocessing. Pharm. Bioprocess. 1, 197–213. 10.4155/pbp.13.18 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

