Quantifying nanoscale forces using machine learning in dynamic atomic force microscopy
- PMID: 35601812
- PMCID: PMC9063738
- DOI: 10.1039/d2na00011c
Quantifying nanoscale forces using machine learning in dynamic atomic force microscopy
Abstract
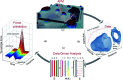
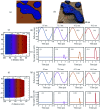
Dynamic atomic force microscopy (AFM) is a key platform that enables topological and nanomechanical characterization of novel materials. This is achieved by linking the nanoscale forces that exist between the AFM tip and the sample to specific mathematical functions through modeling. However, the main challenge in dynamic AFM is to quantify these nanoscale forces without the use of complex models that are routinely used to explain the physics of tip-sample interaction. Here, we make use of machine learning and data science to characterize tip-sample forces purely from experimental data with sub-microsecond resolution. Our machine learning approach is first trained on standard AFM models and then showcased experimentally on a polymer blend of polystyrene (PS) and low density polyethylene (LDPE) sample. Using this algorithm we probe the complex physics of tip-sample contact in polymers, estimate elasticity, and provide insight into energy dissipation during contact. Our study opens a new route in dynamic AFM characterization where machine learning can be combined with experimental methodologies to probe transient processes involved in phase transformation as well as complex chemical and biological phenomena in real-time.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Discrimination of adhesion and viscoelasticity from nanoscale maps of polymer surfaces using bimodal atomic force microscopy.Nanoscale. 2021 Oct 28;13(41):17428-17441. doi: 10.1039/d1nr03437e. Nanoscale. 2021. PMID: 34647552
-
An atomic force microscope tip designed to measure time-varying nanomechanical forces.Nat Nanotechnol. 2007 Aug;2(8):507-14. doi: 10.1038/nnano.2007.226. Epub 2007 Jul 29. Nat Nanotechnol. 2007. PMID: 18654349
-
Different directional energy dissipation of heterogeneous polymers in bimodal atomic force microscopy.RSC Adv. 2019 Sep 2;9(47):27464-27474. doi: 10.1039/c9ra03995c. eCollection 2019 Aug 29. RSC Adv. 2019. PMID: 35529235 Free PMC article.
-
Multiparametric Atomic Force Microscopy Imaging of Biomolecular and Cellular Systems.Acc Chem Res. 2017 Apr 18;50(4):924-931. doi: 10.1021/acs.accounts.6b00638. Epub 2017 Mar 28. Acc Chem Res. 2017. PMID: 28350161 Review.
-
Recent Applications of Advanced Atomic Force Microscopy in Polymer Science: A Review.Polymers (Basel). 2020 May 17;12(5):1142. doi: 10.3390/polym12051142. Polymers (Basel). 2020. PMID: 32429499 Free PMC article. Review.
Cited by
-
Analysis of biofilm assembly by large area automated AFM.NPJ Biofilms Microbiomes. 2025 May 8;11(1):75. doi: 10.1038/s41522-025-00704-y. NPJ Biofilms Microbiomes. 2025. PMID: 40341406 Free PMC article.
-
High-Throughput Nanorheology of Living Cells Powered by Supervised Machine Learning.Adv Intell Syst. 2025 Aug;7(8):2400867. doi: 10.1002/aisy.202400867. Epub 2025 Apr 15. Adv Intell Syst. 2025. PMID: 40852088 Free PMC article.
-
Nonlinear Harmonics: A Gateway to Enhanced Image Contrast and Material Discrimination.Adv Sci (Weinh). 2025 Mar;12(11):e2411556. doi: 10.1002/advs.202411556. Epub 2025 Jan 28. Adv Sci (Weinh). 2025. PMID: 39876697 Free PMC article.
-
Characteristics and Functionality of Cantilevers and Scanners in Atomic Force Microscopy.Materials (Basel). 2023 Sep 24;16(19):6379. doi: 10.3390/ma16196379. Materials (Basel). 2023. PMID: 37834515 Free PMC article. Review.
-
Machine learning-augmented surface-enhanced spectroscopy toward next-generation molecular diagnostics.Nanoscale Adv. 2022 Nov 7;5(3):538-570. doi: 10.1039/d2na00608a. eCollection 2023 Jan 31. Nanoscale Adv. 2022. PMID: 36756499 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

