Bacterial Outer Membrane Permeability Increase Underlies the Bactericidal Effect of Fatty Acids From Hermetia illucens (Black Soldier Fly) Larvae Fat Against Hypermucoviscous Isolates of Klebsiella pneumoniae
- PMID: 35602017
- PMCID: PMC9121012
- DOI: 10.3389/fmicb.2022.844811
Bacterial Outer Membrane Permeability Increase Underlies the Bactericidal Effect of Fatty Acids From Hermetia illucens (Black Soldier Fly) Larvae Fat Against Hypermucoviscous Isolates of Klebsiella pneumoniae
Abstract
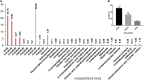
Behind expensive treatments, Klebsiella pneumoniae infections account for extended hospitalization's high mortality rates. This study aimed to evaluate the activity and mechanism of the antimicrobial action of a fatty acid-containing extract (AWME3) isolated from Hermetia illucens (HI) larvae fat against K. pneumoniae subsp. pneumoniae standard NDM-1 carbapenemase-producing ATCC BAA-2473 strain, along with a wild-type hypermucoviscous clinical isolate, strain K. pneumoniae subsp. pneumoniae KPi1627, and an environmental isolate, strain K. pneumoniae subsp. pneumoniae KPM9. We classified these strains as extensive multidrug-resistant (XDR) or multiple antibiotic-resistant (MDR) demonstrated by a susceptibility assay against 14 antibiotics belonging to ten classes of antibiotics. Antibacterial properties of fatty acids extracted from the HI larvae fat were evaluated using disk diffusion method, microdilution, minimum inhibitory concentration (MIC), minimum bactericidal concentration (MBC), half of the inhibitory concentration (MIC50), and bactericidal assays. In addition, the cytotoxocity of AWME3 was tested on human HEK293 cells, and AWME3 lipid profile was determined by gas chromatography-mass spectrometry (GC-MS) analysis. For the first time, we demonstrated that the inhibition zone diameter (IZD) of fatty acid-containing extract (AWME3) of the HI larvae fat tested at 20 mg/ml was 16.52 ± 0.74 and 14.23 ± 0.35 mm against colistin-resistant KPi1627 and KPM9, respectively. It was 19.72 ± 0.51 mm against the colistin-susceptible K. pneumoniae ATCC BAA-2473 strain. The MIC and MBC were 250 μg/ml for all the tested bacteria strains, indicating the bactericidal effect of AWME3. The MIC50 values were 155.6 ± 0.009 and 160.1 ± 0.008 μg/ml against the KPi1627 and KPM9 isolates, respectively, and 149.5 ± 0.013 μg/ml against the ATCC BAA-2473 strain in the micro-dilution assay. For the first time, we demonstrated that AWME3 dose-dependently increased bacterial cell membrane permeability as determined by the relative electric conductivity (REC) of the K. pneumoniae ATCC BAA-2473 suspension, and that none of the strains did not build up resistance to extended AWME3 treatment using the antibiotic resistance assay. Cytotoxicity assay showed that AWME3 is safe for human HEK293 cells at IC50 266.1 μg/ml, while bactericidal for all the strains of bacteria at the same concentration. Free fatty acids (FFAs) and their derivatives were the significant substances among 33 compounds identified by the GC-MS analysis of AWME3. Cis-oleic and palmitoleic acids represent the most abundant unsaturated FAs (UFAs), while palmitic, lauric, stearic, and myristic acids were the most abundant saturated FAs (SFAs) of the AWME3 content. Bactericidal resistant-free AWM3 mechanism of action provides a rationale interpretations and the utility of HI larvae fat to develop natural biocidal resistance-free formulations that might be promising therapeutic against Gram-negative MDR bacteria causing nosocomial infections.
Keywords: H. illucens; Klebsiella pneumoniae; MDR bacteria; NDM-1 carbapenemase-producing Enterobacteriaceae (CPE); colistin; free fatty acids; hypermucoviscous; nosocomial isolates.
Copyright © 2022 Mohamed, Marusich, Afanasev and Leonov.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
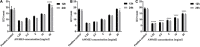



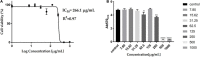
Similar articles
-
A unique combination of natural fatty acids from Hermetia illucens fly larvae fat effectively combats virulence factors and biofilms of MDR hypervirulent mucoviscus Klebsiella pneumoniae strains by increasing Lewis acid-base/van der Waals interactions in bacterial wall membranes.Front Cell Infect Microbiol. 2024 Jul 25;14:1408179. doi: 10.3389/fcimb.2024.1408179. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39119288 Free PMC article.
-
Fatty Acids-Enriched Fractions of Hermetia illucens (Black Soldier Fly) Larvae Fat Can Combat MDR Pathogenic Fish Bacteria Aeromonas spp.Int J Mol Sci. 2021 Aug 17;22(16):8829. doi: 10.3390/ijms22168829. Int J Mol Sci. 2021. PMID: 34445533 Free PMC article.
-
In vitro and in vivo bactericidal activity of ceftazidime-avibactam against Carbapenemase-producing Klebsiella pneumoniae.Antimicrob Resist Infect Control. 2018 Nov 21;7:142. doi: 10.1186/s13756-018-0435-9. eCollection 2018. Antimicrob Resist Infect Control. 2018. PMID: 30479755 Free PMC article.
-
A Molecular Perspective on Colistin and Klebsiella pneumoniae: Mode of Action, Resistance Genetics, and Phenotypic Susceptibility.Diagnostics (Basel). 2021 Jun 25;11(7):1165. doi: 10.3390/diagnostics11071165. Diagnostics (Basel). 2021. PMID: 34202395 Free PMC article. Review.
-
In vivo and in vitro inhibition effect of propolis on Klebsiella pneumoniae: A review.Ann Med Surg (Lond). 2022 Aug 18;81:104388. doi: 10.1016/j.amsu.2022.104388. eCollection 2022 Sep. Ann Med Surg (Lond). 2022. PMID: 36147103 Free PMC article. Review.
Cited by
-
A Sustainable Synthesis of Novel 2-(3,4-Disubstituted phenyl)benzoxazole Derivatives and Their Antiproliferative and Antibacterial Evaluation.Molecules. 2025 Apr 15;30(8):1767. doi: 10.3390/molecules30081767. Molecules. 2025. PMID: 40333762 Free PMC article.
-
Antibacterial and Antifungal Potential of Hermetia illucens Hemolymph Contained-Peptides.Probiotics Antimicrob Proteins. 2025 Aug 7. doi: 10.1007/s12602-025-10697-x. Online ahead of print. Probiotics Antimicrob Proteins. 2025. PMID: 40773039
-
Exploring the intricate studies on low-density polyethylene (LDPE) biodegradation by Bacillus cereus AP-01, isolated from the gut of Styrofoam-fed Tenebrio molitor larvae.Biodegradation. 2025 Jan 7;36(1):12. doi: 10.1007/s10532-024-10107-z. Biodegradation. 2025. PMID: 39775270
-
The Variety of Applications of Hermetia illucens in Industrial and Agricultural Areas-Review.Biology (Basel). 2022 Dec 22;12(1):25. doi: 10.3390/biology12010025. Biology (Basel). 2022. PMID: 36671718 Free PMC article. Review.
-
A unique combination of natural fatty acids from Hermetia illucens fly larvae fat effectively combats virulence factors and biofilms of MDR hypervirulent mucoviscus Klebsiella pneumoniae strains by increasing Lewis acid-base/van der Waals interactions in bacterial wall membranes.Front Cell Infect Microbiol. 2024 Jul 25;14:1408179. doi: 10.3389/fcimb.2024.1408179. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39119288 Free PMC article.
References
-
- Agoramoorthy G., Chandrasekaran M., Venkatesalu V., Hsu M. J. (2007). Antibacterial and antifungal activities of fatty acid methyl esters of the blind-your-eye mangrove from India. Braz. J. Microbiol. 38 739–742. 10.1590/S1517-83822007000400028 - DOI
-
- Algammal A. M., Hashem H. R., Alfifi K. J., Hetta H. F., Sheraba N. S., Ramadan H., et al. (2021). atpD gene sequencing, multidrug resistance traits, virulence-determinants, and antimicrobial resistance genes of emerging XDR and MDR-Proteus mirabilis. Sci. Rep. 11:9476. 10.1038/s41598-021-88861-w - DOI - PMC - PubMed
-
- Algammal A. M., Hetta H. F., Batiha G. E., Hozzein W. N., El Kazzaz W. M., Hashem H. R., et al. (2020a). Virulence-determinants and antibiotic-resistance genes of MDR-E. coli isolated from secondary infections following FMD-outbreak in cattle. Sci. Rep. 10:19779. 10.1038/s41598-020-75914-9 - DOI - PMC - PubMed
-
- Algammal A. M., Hetta H. F., Elkelish A., Alkhalifah D. H. H., Hozzein W. N., Batiha G. E. S., et al. (2020b). Methicillin-resistant staphylococcus aureus (MRSA): one health perspective approach to the bacterium epidemiology, virulence factors, antibiotic-resistance, and zoonotic impact. Infect. Drug Resist. 13 3255–3265. 10.2147/IDR.S272733 - DOI - PMC - PubMed
-
- Algammal A. M., Mabrok M., Sivaramasamy E., Youssef F. M., Atwa M. H., El-kholy A. W., et al. (2020c). Emerging MDR-Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and bla TEM, bla CTX-M, and tetA antibiotic-resistance genes. Sci. Rep. 10:15961. 10.1038/s41598-020-72264-4 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

