Insight Into Distinct Functional Roles of the Flagellar ATPase Complex for Flagellar Assembly in Salmonella
- PMID: 35602071
- PMCID: PMC9114704
- DOI: 10.3389/fmicb.2022.864178
Insight Into Distinct Functional Roles of the Flagellar ATPase Complex for Flagellar Assembly in Salmonella
Abstract
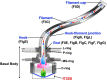
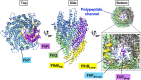
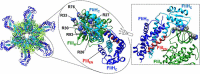
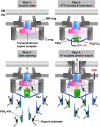
Most motile bacteria utilize the flagellar type III secretion system (fT3SS) to construct the flagellum, which is a supramolecular motility machine consisting of basal body rings and an axial structure. Each axial protein is translocated via the fT3SS across the cytoplasmic membrane, diffuses down the central channel of the growing flagellar structure and assembles at the distal end. The fT3SS consists of a transmembrane export complex and a cytoplasmic ATPase ring complex with a stoichiometry of 12 FliH, 6 FliI and 1 FliJ. This complex is structurally similar to the cytoplasmic part of the FOF1 ATP synthase. The export complex requires the FliH12-FliI6-FliJ1 ring complex to serve as an active protein transporter. The FliI6 ring has six catalytic sites and hydrolyzes ATP at an interface between FliI subunits. FliJ binds to the center of the FliI6 ring and acts as the central stalk to activate the export complex. The FliH dimer binds to the N-terminal domain of each of the six FliI subunits and anchors the FliI6-FliJ1 ring to the base of the flagellum. In addition, FliI exists as a hetero-trimer with the FliH dimer in the cytoplasm. The rapid association-dissociation cycle of this hetero-trimer with the docking platform of the export complex promotes sequential transfer of export substrates from the cytoplasm to the export gate for high-speed protein transport. In this article, we review our current understanding of multiple roles played by the flagellar cytoplasmic ATPase complex during efficient flagellar assembly.
Keywords: ATPase; F0F1 ATP synthase; bacterial flagella; flagellar assembly; protein translocation; proton motive force (pmf); type III secretion system (T3SS).
Copyright © 2022 Minamino, Kinoshita and Namba.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources

