Patient-derived organoids reflect the genetic profile of endometrial tumors and predict patient prognosis
- PMID: 35602206
- PMCID: PMC9053236
- DOI: 10.1038/s43856-021-00019-x
Patient-derived organoids reflect the genetic profile of endometrial tumors and predict patient prognosis
Erratum in
-
Erratum: Author Correction: Patient-derived organoids reflect the genetic profile of endometrial tumors and predict patient prognosis.Commun Med (Lond). 2021 Aug 10;1:22. doi: 10.1038/s43856-021-00022-2. eCollection 2021. Commun Med (Lond). 2021. PMID: 35604807 Free PMC article.
Abstract
Background: A major hurdle in translational endometrial cancer (EC) research is the lack of robust preclinical models that capture both inter- and intra-tumor heterogeneity. This has hampered the development of new treatment strategies for people with EC.
Methods: EC organoids were derived from resected patient tumor tissue and expanded in a chemically defined medium. Established EC organoids were orthotopically implanted into female NSG mice. Patient tissue and corresponding models were characterized by morphological evaluation, biomarker and gene expression and by whole exome sequencing. A gene signature was defined and its prognostic value was assessed in multiple EC cohorts using Mantel-Cox (log-rank) test. Response to carboplatin and/or paclitaxel was measured in vitro and evaluated in vivo. Statistical difference between groups was calculated using paired t-test.
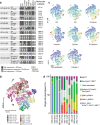
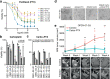
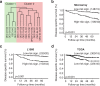
Results: We report EC organoids established from EC patient tissue, and orthotopic organoid-based patient-derived xenograft models (O-PDXs). The EC organoids and O-PDX models mimic the tissue architecture, protein biomarker expression and genetic profile of the original tissue. Organoids show heterogenous sensitivity to conventional chemotherapy, and drug response is reproduced in vivo. The relevance of these models is further supported by the identification of an organoid-derived prognostic gene signature. This signature is validated as prognostic both in our local patient cohorts and in the TCGA endometrial cancer cohort.
Conclusions: We establish robust model systems that capture both the diversity of endometrial tumors and intra-tumor heterogeneity. These models are highly relevant preclinical tools for the elucidation of the molecular pathogenesis of EC and identification of potential treatment strategies.
Keywords: Cancer genomics; Cancer models; Chemotherapy; Endometrial cancer.
© The Author(s) 2021, corrected publication 2021.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures



References
-
- Carcangiu, M. L. WHO classification of tumours of female reproductive organs, fourth edition. Lyon: International Agency for Research on Cancer (I A R C)(UN) (2014).
-
- Concin, N. et al. ESGO/ESTRO/ESP Guidelines for the management of patients with endometrial carcinoma. Int J Gynecol Cancer31, 12–39 (2020). - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

