Model-based estimation of transmissibility and reinfection of SARS-CoV-2 P.1 variant
- PMID: 35602219
- PMCID: PMC9053218
- DOI: 10.1038/s43856-021-00048-6
Model-based estimation of transmissibility and reinfection of SARS-CoV-2 P.1 variant
Abstract
Background: The SARS-CoV-2 variant of concern (VOC) P.1 (Gamma variant) emerged in the Amazonas State, Brazil, in November 2020. The epidemiological consequences of its mutations have not been widely studied, despite detection of P.1 in 36 countries, with local transmission in at least 5 countries. A range of mutations are seen in P.1, ten of them in the spike protein. It shares mutations with VOCs previously detected in the United Kingdom (B.1.1.7, Alpha variant) and South Africa (B.1.351, Beta variant).
Methods: We estimated the transmissibility and reinfection of P.1 using a model-based approach, fitting data from the national health surveillance of hospitalized individuals and frequency of the P.1 variant in Manaus from December-2020 to February-2021.
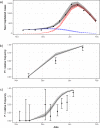
Results: Here we estimate that the new variant is about 2.6 times more transmissible (95% Confidence Interval: 2.4-2.8) than previous circulating variant(s). Manaus already had a high prevalence of individuals previously affected by the SARS-CoV-2 virus and our fitted model attributed 28% of Manaus cases in the period to reinfections by P.1, confirming the importance of reinfection by this variant. This value is in line with estimates from blood donors samples in Manaus city.
Conclusions: Our estimates rank P.1 as one of the most transmissible among the SARS-CoV-2 VOCs currently identified, and potentially as transmissible as the posteriorly detected VOC B.1.617.2 (Delta variant), posing a serious threat and requiring measures to control its global spread.
Keywords: Dynamical systems; Viral infection.
© The Author(s) 2021.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures
References
-
- NIID Japan. Brief report: New variant strain of sars-cov-2 identified in travelers from Brazil. coronavirus disease 4. https://www.niid.go.jp/niid/en/2019-ncov-e/10108-covid19-33-en.html (2021).
-
- Naveca, F. et al. Phylogenetic relationship of sars-cov-2 sequences from Amazonas with emerging Brazilian variants harboring mutations E484K and N501Y in the spike protein. Virological.org. https://virological.org/t/phylogenetic-relationship-of-sars-cov-2-sequen... (2021).
-
- Naveca, F. et al. COVID-19 in Amazonas, Brazil, was driven by the persistence of endemic lineages and P.1 emergence. Nat. Med. https://www.nature.com/articles/s41591-021-01378-7 (2021). - PubMed
-
- Davies, N. G. et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. https://science.sciencemag.org/content/early/2021/03/03/science.abg3055. https://science.sciencemag.org/content/early/2021/03/03/science.abg3055.... (2021). - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

