Increased glycine contributes to synaptic dysfunction and early mortality in Nprl2 seizure model
- PMID: 35602938
- PMCID: PMC9118754
- DOI: 10.1016/j.isci.2022.104334
Increased glycine contributes to synaptic dysfunction and early mortality in Nprl2 seizure model
Abstract
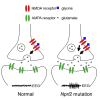
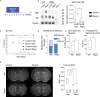
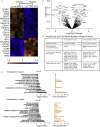
Targeted therapies for epilepsies associated with the mTORC1 signaling negative regulator GATOR1 are lacking. NPRL2 is a subunit of the GATOR1 complex and mutations in GATOR1 subunits, including NPRL2, are associated with epilepsy. To delineate the mechanisms underlying NPRL2-related epilepsies, we created a mouse (Mus musculus) model with neocortical loss of Nprl2. Mutant mice have increased mTORC1 signaling and exhibit spontaneous seizures. They also display abnormal synaptic function characterized by increased evoked and spontaneous EPSC and decreased evoked and spontaneous IPSC frequencies, respectively. Proteomic and metabolomics studies of Nprl2 mutants revealed alterations in known epilepsy-implicated proteins and metabolic pathways, including increases in the neurotransmitter, glycine. Furthermore, glycine actions on the NMDA receptor contribute to the electrophysiological and survival phenotypes of these mice. Taken together, in this neuronal Nprl2 model, we delineate underlying molecular, metabolic, and electrophysiological mechanisms contributing to mTORC1-related epilepsy, providing potential therapeutic targets for epilepsy.
Keywords: metabolomics; molecular neuroscience; neuroscience; omics.
© 2022 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

